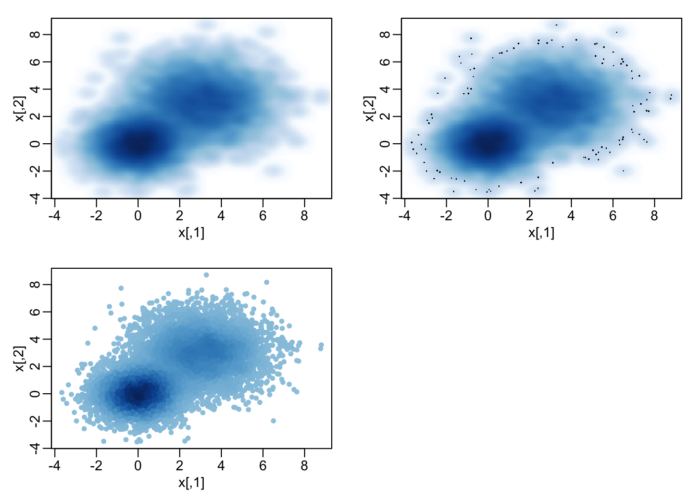
- ## A largish data set
- n <- 10000
- x1 <- matrix(rnorm(n), ncol = 2)
- x2 <- matrix(rnorm(n, mean = 3, sd = 1.5), ncol = 2)
- x <- rbind(x1, x2)
-
- oldpar <- par(mfrow = c(2, 2), mar=.1+c(3,3,1,1), mgp = c(1.5, 0.5, 0))
- smoothScatter(x, nrpoints = 0)
- smoothScatter(x)
-
- ## but considerably *less* efficient for really large data:
- plot(x, col = densCols(x), pch = 20)
-
- ## use with pairs:
- par(mfrow = c(1, 1))
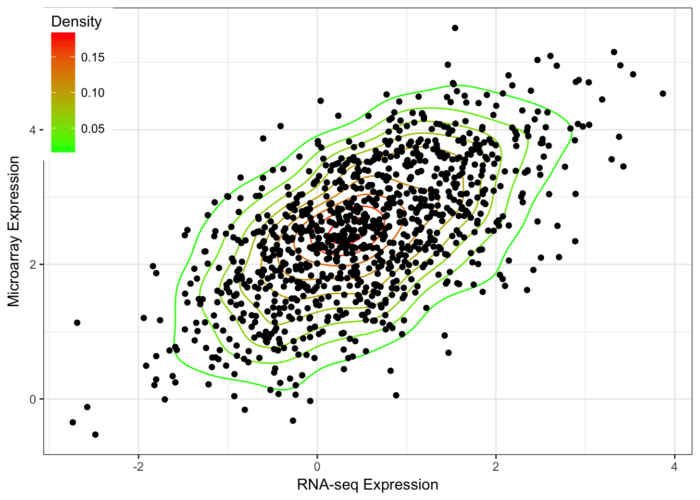
- library(MASS)
- library(ggplot2)
- ## Warning: package 'ggplot2' was built under R version 3.2.5
- n <- 1000
- x <- mvrnorm(n, mu=c(.5,2.5), Sigma=matrix(c(1,.6,.6,1), ncol=2))
- df = data.frame(x); colnames(df) = c("x","y")
-
- commonTheme = list(labs(color="Density",fill="Density",
- x="RNA-seq Expression",
- y="Microarray Expression"),
- theme_bw(),
- theme(legend.position=c(0,1),
- legend.justification=c(0,1)))
-
- ggplot(data=df,aes(x,y)) +
- geom_density2d(aes(colour=..level..)) +
- scale_colour_gradient(low="green",high="red") +
- geom_point() + commonTheme
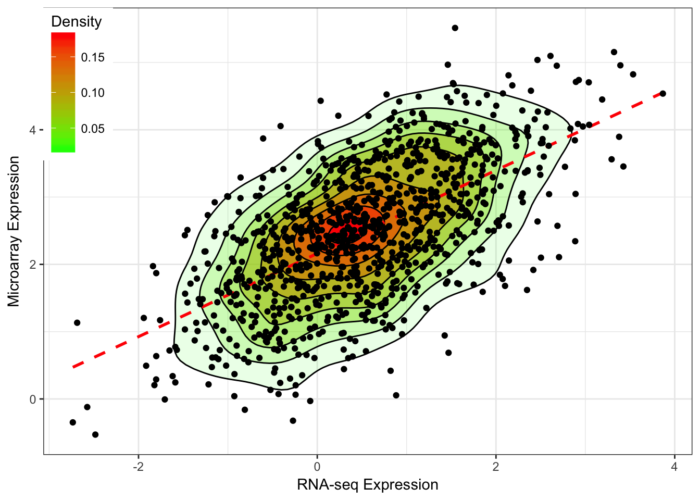
- ggplot(data=df,aes(x,y)) +
- stat_density2d(aes(fill=..level..,alpha=..level..),geom='polygon',colour='black') +
- scale_fill_continuous(low="green",high="red") +
- geom_smooth(method=lm,linetype=2,colour="red",se=F) +
- guides(alpha="none") +
- geom_point() + commonTheme
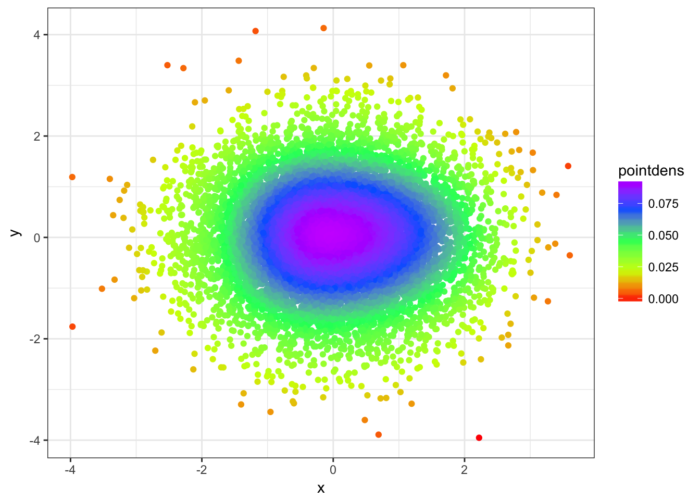
- n <- 10000
- x <- rnorm(n)
- y <- rnorm(n)
- DF <- data.frame(x,y)
- library(LSD)
- heatscatter(DF[,1],DF[,2])
- # generare random data, swap this for yours :-)!
- n <- 10000
- x <- rnorm(n)
- y <- rnorm(n)
- DF <- data.frame(x,y)
-
- # Calculate 2d density over a grid
- library(MASS)
- dens <- kde2d(x,y)
-
- # create a new data frame of that 2d density grid
- # (needs checking that I haven't stuffed up the order here of z?)
- gr <- data.frame(with(dens, expand.grid(x,y)), as.vector(dens$z))
- names(gr) <- c("xgr", "ygr", "zgr")
-
- # Fit a model
- mod <- loess(zgr~xgr*ygr, data=gr)
-
- # Apply the model to the original data to estimate density at that point
- DF$pointdens <- predict(mod, newdata=data.frame(xgr=x, ygr=y))
-
- # Draw plot
- library(ggplot2)
- ggplot(DF, aes(x=x,y=y, color=pointdens)) + geom_point() + scale_colour_gradientn(colours = rainbow(5)) + theme_bw()