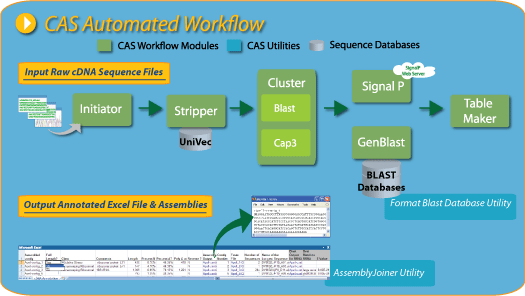
The Desktop cDNA Annotation System (dCAS) automates large-scale cDNA sequence analysis. dCAS allows you to import raw cDNA sequences, clean sequences, build sequence contigs, perform SignalP analysis, BLAST contigs against numerous BLAST databases, and view the results in a Microsoft Excel worksheet. Manual curation of each gene can be performed in the excel file and then easily re-imported into the cDNA Annotation System. dCAS enables you to view, justify, and edit the 6-frame peptide translation. As a final step in the workflow, annotated genes and their corresponding translated products can be written into FASTA files for submission to GenBank.
相关链接:http://exon.niaid.nih.gov/cas/