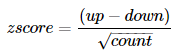GOplot 包通过封装好的函数可视化基因功能分析结果
#1. 安装
- > install.packages('GOplot')
#2. GOplot 内置数据
##2.1 脑和心脏内皮细胞的转录组数据
Form paper: Nolan et al. 2013,GEO accession: GSE47067.
| Name | Description | Dimension |
|---|---|---|
| EC$eset | Data frame of normalized expression values of brain and heart endothelial cells (3 replicates) | 20644 x 7 |
| EC$genelist | Data frame of differentially expressed genes (adjusted p-value < 0.05) | 2039 x 7 |
| EC$david | Data frame of results from a functional analysis of the differentially expressed genes performed with DAVID | 174 x 5 |
| EC$genes | Data frame of selected genes with logFC | 37 x 2 |
| EC$process | Character vector of selected enriched biological processes | 7 |
##2.2 查看内置数据格式
导入数据
- > library(GOplot)
- > data(EC)
基因富集结果查看
- > head(EC$david,2)
- Category ID Term
- 1 BP GO:0007507 heart development
- 2 BP GO:0001944 vasculature development
- Genes
- 1 DLC1, NRP2, NRP1, EDN1, PDLIM3, GJA1, TTN, GJA5, ZIC3, TGFB2, CERKL, GATA6, COL4A3BP, GAB1, SEMA3C, MKL2, SLC22A5, MB, PTPRJ, RXRA, VANGL2, MYH6, TNNT2, HHEX, MURC, MIB1, FOXC2, FOXC1, ADAM19, MYL2, TCAP, EGLN1, SOX9, ITGB1, CHD7, HEXIM1, PKD2, NFATC4, PCSK5, ACTC1, TGFBR2, NF1, HSPG2, SMAD3, TBX1, TNNI3, CSRP3, FOXP1, KCNJ8, PLN, TSC2, ATP6V0A1, TGFBR3, HDAC9
- 2 GNA13, ACVRL1, NRP1, PGF, IL18, LEPR, EDN1, GJA1, FOXO1, GJA5, TGFB2, WARS, CERKL, APOE, CXCR4, ANG, SEMA3C, NOS2, MKL2, FGF2, RAPGEF1, PTPRJ, RECK, EFNB2, VASH1, PNPLA6, THY1, MIB1, NUS1, FOXC2, FOXC1, CAV1, CDH2, MEIS1, WT1, CDH5, PTK2, FBXW8, CHD7, PLCD1, PLXND1, FIGF, PPAP2B, MAP2K1, TBX4, TGFBR2, NF1, TBX1, TNNI3, LAMA4, MEOX2, ECSCR, HBEGF, AMOT, TGFBR3, HDAC7
- adj_pval
- 1 2.17e-06
- 2 1.04e-05
查看选择的基因
- > head(EC$genelist)
- ## ID logFC AveExpr t P.Value adj.P.Val B
- ## 1 Slco1a4 6.645388 1.2168670 88.65515 1.32e-18 2.73e-14 29.02715
- ## 2 Slc19a3 6.281525 1.1600468 69.95094 2.41e-17 2.49e-13 27.62917
- ## 3 Ddc 4.483338 0.8365231 65.57836 5.31e-17 3.65e-13 27.18476
- ## 4 Slco1c1 6.469384 1.3558865 59.87613 1.62e-16 8.34e-13 26.51242
- ## 5 Sema3c 5.515630 2.3252117 58.53141 2.14e-16 8.81e-13 26.33626
- ## 6 Slc38a3 4.761755 0.9218670 54.11559 5.58e-16 1.76e-12 25.70308
构建画图数据:circle_dat()
- > circ <- circle_dat(EC$david, EC$genelist)
- > head(circ)
- category ID term count genes logFC adj_pval zscore
- BP GO:0007507 heart development 54 DLC1 -0.9707875 2.17e-06 -0.8164966
- BP GO:0007507 heart development 54 NRP2 -1.5153173 2.17e-06 -0.8164966
- BP GO:0007507 heart development 54 NRP1 -1.1412315 2.17e-06 -0.8164966
- BP GO:0007507 heart development 54 EDN1 1.3813006 2.17e-06 -0.8164966
- BP GO:0007507 heart development 54 PDLIM3 -0.8876939 2.17e-06 -0.8164966
- BP GO:0007507 heart development 54 GJA1 -0.8179480 2.17e-06 -0.8164966
zscore: 每个GO term下上调(logFC>0)基因数和下调基因数的差与注释到GO term基因数平方根的商。
#3. 画图
##3.1 条形图(GOBar())
画BP下的GO term
- > GOBar(subset(circ, category == 'BP')
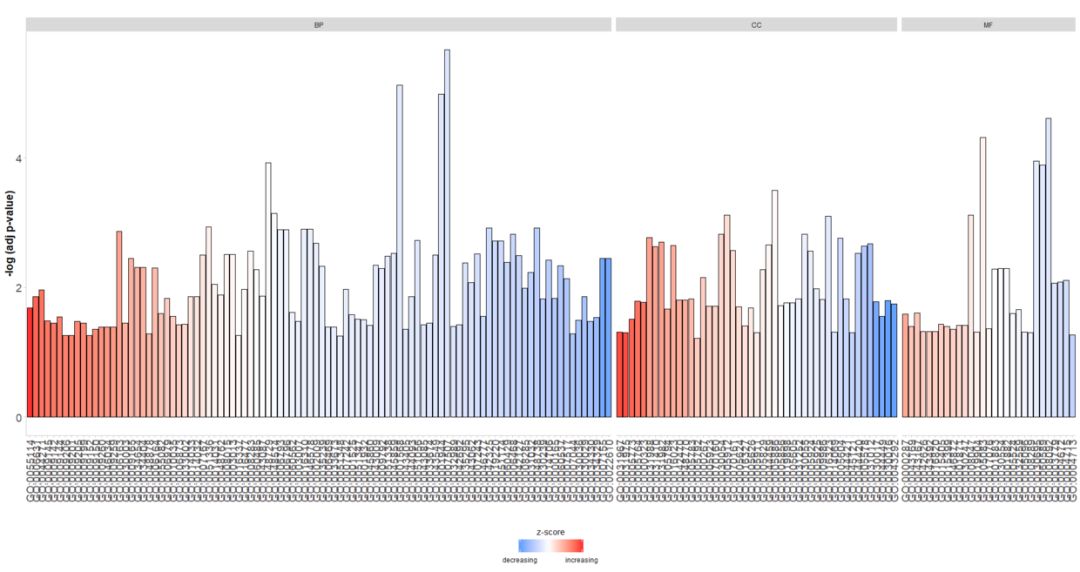
分面同时展示BP, CC, MF的GO term
- > GOBar(circ, display = 'multiple')
#3.2 气泡图(GOBubble())
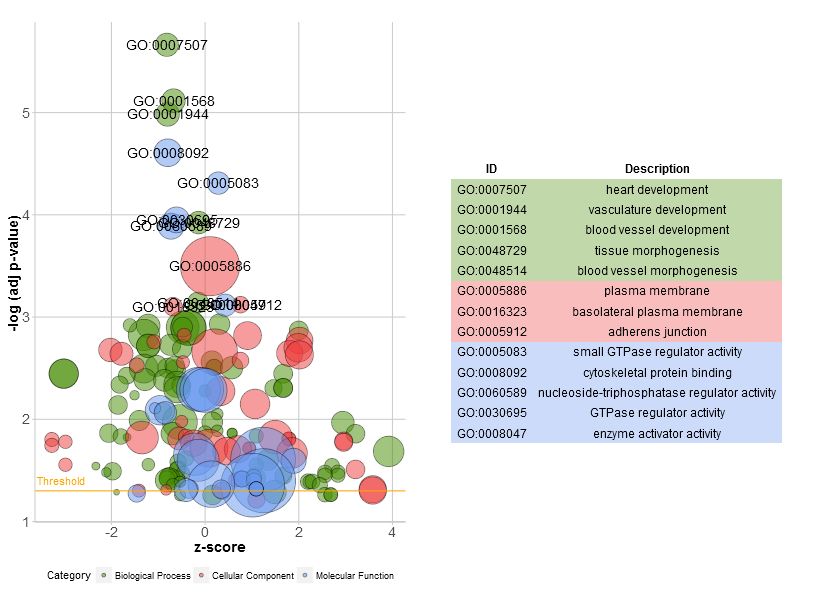
- > GOBubble(circ, labels = 3)
上图中:X轴是z-score; Y轴是多重矫正后p值的负对数;圈大小展示GO Term下基因数。
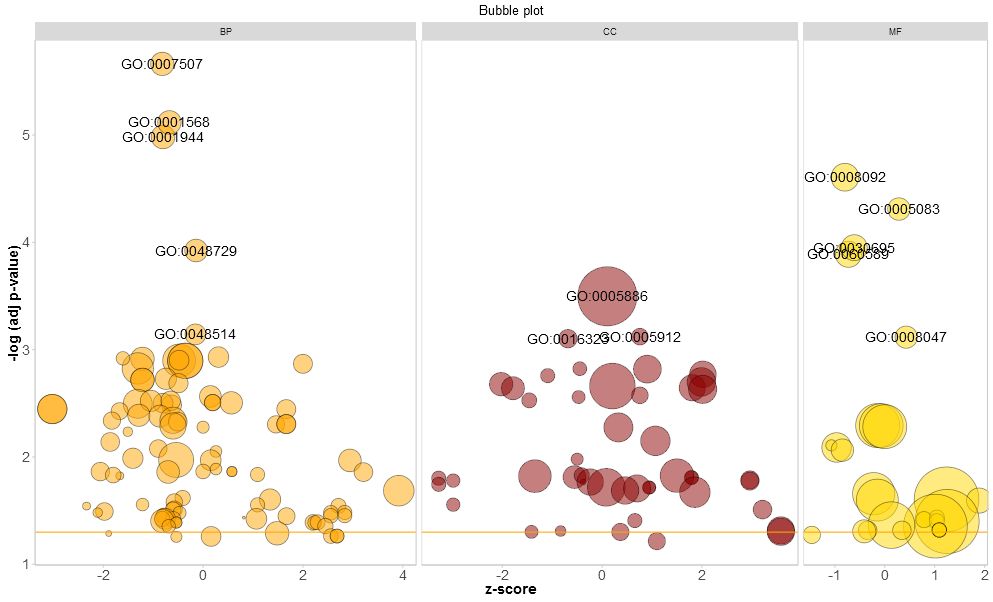
分面同时展示BP, CC, MF的气泡图
- > GOBubble(circ, title = 'Bubble plot', colour = c('orange', 'darkred', 'gold'), display = 'multiple', labels = 3)
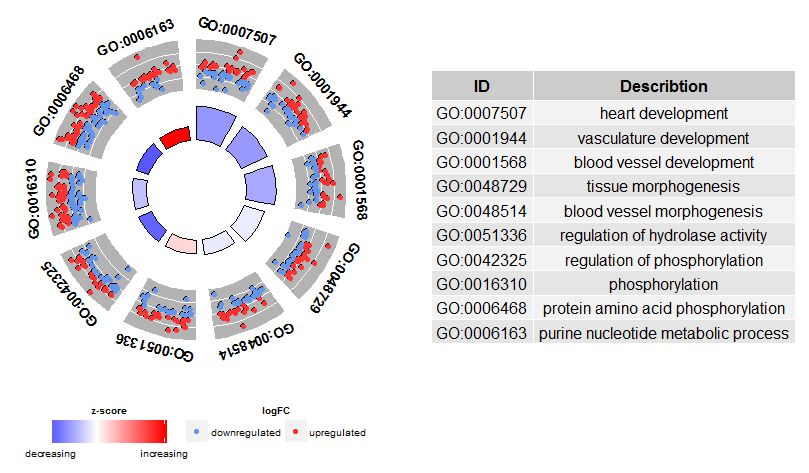
#2.3 圈图展示基因富集分析结果(GOCircle())
- > GOCircle(circ)
默认展示circ 数据前10个GO Term,通过参数nsub调整需要展示的GO Term
根据GO Term选择要展示的GO Term
- > GOCircle(circ, nsub = c('GO:0007507', 'GO:0001568', 'GO:0001944', 'GO:0048729', 'GO:0048514', 'GO:0005886', 'GO:0008092', 'GO:0008047'))
选择要展示的GO Term数量
- GOCircle(circ, nsub = 10)
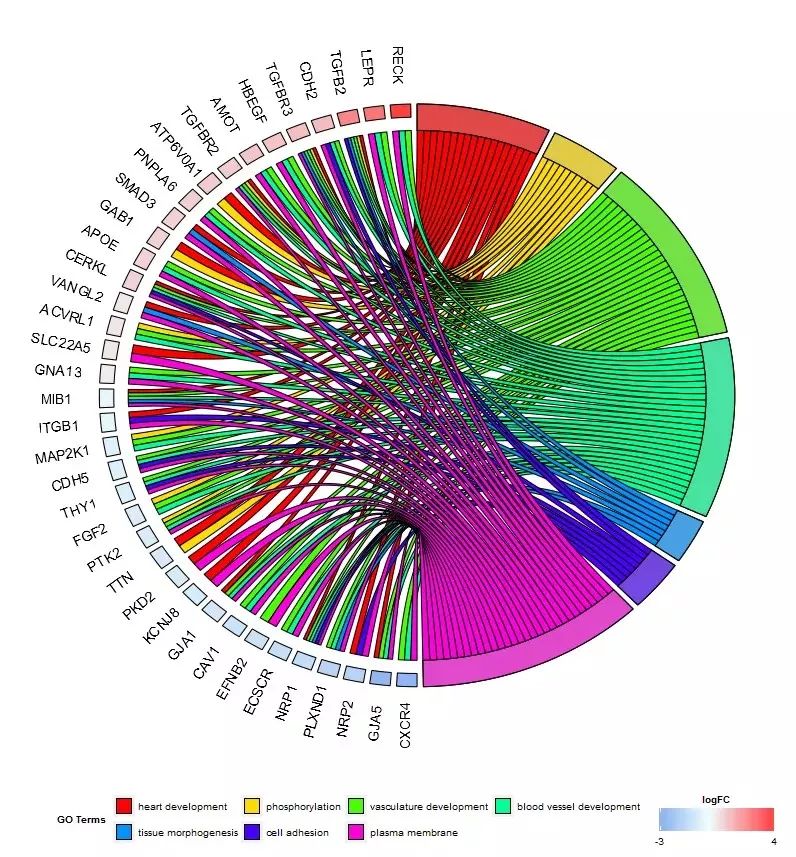
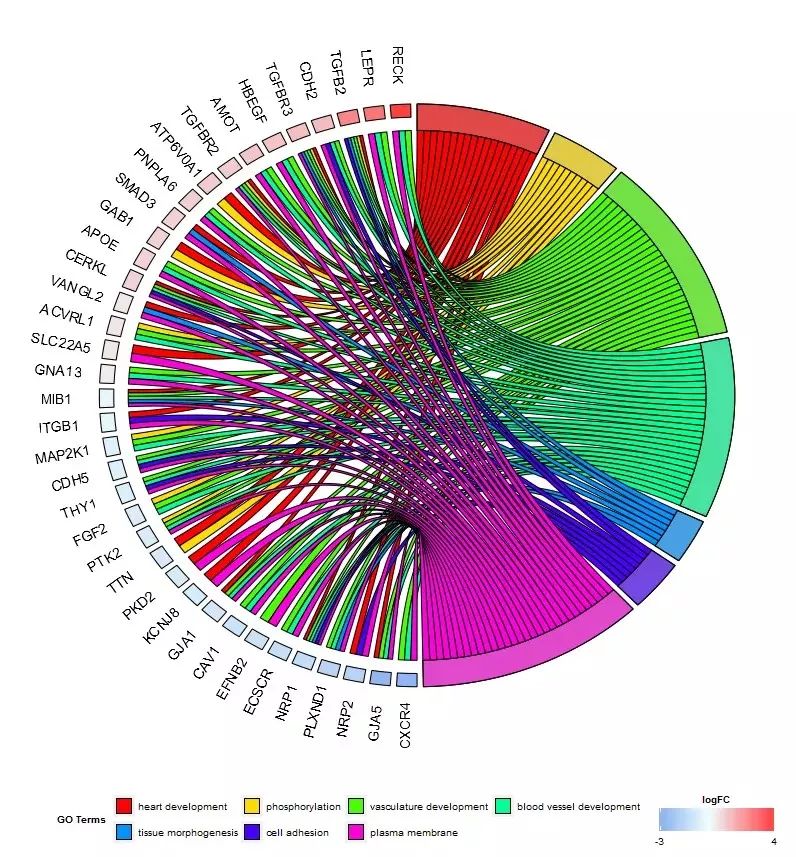
#2.4 展示基因与GO Terms关系的圈图 (GOChord())
chord_dat ()将作图数据构建成GOChord() 要求的输入格式;一个二进制的关系矩阵,1表示基因属于该GO Term,0与之相反。
选择感兴趣的基因
- > head(EC$genes)
- ## ID logFC
- ## 1 PTK2 -0.6527904
- ## 2 GNA13 0.3711599
- ## 3 LEPR 2.6539788
- ## 4 APOE 0.8698346
- ## 5 CXCR4 -2.5647537
- ## 6 RECK 3.6926860
选择感兴趣的GO Term
- > EC$process
- ## [1] "heart development" "phosphorylation"
- ## [3] "vasculature development" "blood vessel development"
- ## [5] "tissue morphogenesis" "cell adhesion"
- ## [7] "plasma membrane"
构建画图数据
- #chord_dat(data, genes, process)
- #genes、process其中任何一个参数不指定,默认使用对应的全部数据
- > chord <- chord_dat(circ, EC$genes, EC$process)
- > head(chord)
- ## heart development phosphorylation vasculature development
- ## PTK2 0 1 1
- ## GNA13 0 0 1
- ## LEPR 0 0 1
- ## APOE 0 0 1
- ## CXCR4 0 0 1
- ## RECK 0 0 1
- ## blood vessel development tissue morphogenesis cell adhesion
- ## PTK2 1 0 0
- ## GNA13 1 0 0
- ## LEPR 1 0 0
- ## APOE 1 0 0
- ## CXCR4 1 0 0
- ## RECK 1 0 0
- ## plasma membrane logFC
- ## PTK2 1 -0.6527904
- ## GNA13 1 0.3711599
- ## LEPR 1 2.6539788
- ## APOE 1 0.8698346
- ## CXCR4 1 -2.5647537
- ## RECK 1 3.6926860
画图
- chord <- chord_dat(data = circ, genes = EC$genes, process = EC$process)
GOChord(chord, space = 0.02, gene.order = 'logFC', gene.space = 0.25, gene.size = 5)
GOChord() 参数
- GOChord(data, title, space, gene.order, gene.size, gene.space, nlfc = 1,
- lfc.col, lfc.min, lfc.max, ribbon.col, border.size, process.label, limit)
- #data: 二进制矩阵
- #title:标题
- #space:基因对应方块之间的距离
- #gene.order:基因排列顺序
- #gene.size:基因标签大小
- #nlfc:logFC 列的数目
- #lfc.col:LFC颜色,定义模式:c(color for low values, color for the mid point, color for the high values)
- #lfc.min:LFC最小值
- #lfc.max:LFC最大值
- #ribbon.col:向量定义基因与GO Term间条带颜色
- #border.size:基因与GO Term间条带边框粗细
- #process.label:GO Term 图例文字大小
- #limit:c(3, 2),两个数字;第一个参数筛选基因(保留至少存在于3个GO Term的基因),第二个参数筛选GO Term(保留至少包含2个基因的GO Term )
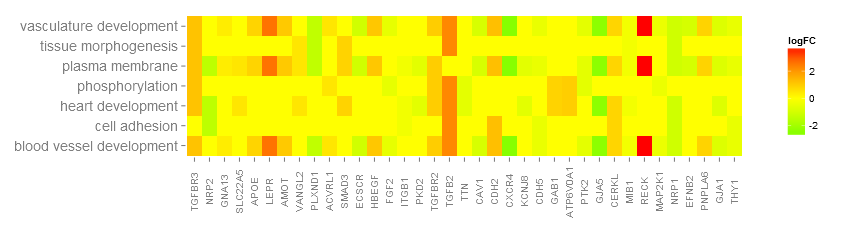
#3.5 基因与GO Term的热图(GOHeat)
nlfc = 1:颜色对应logFC nlfc = 0:颜色对应每个基因注释了到了几个GO Term
- > GOHeat(chord, nlfc = 1, fill.col = c('red', 'yellow', 'green'))
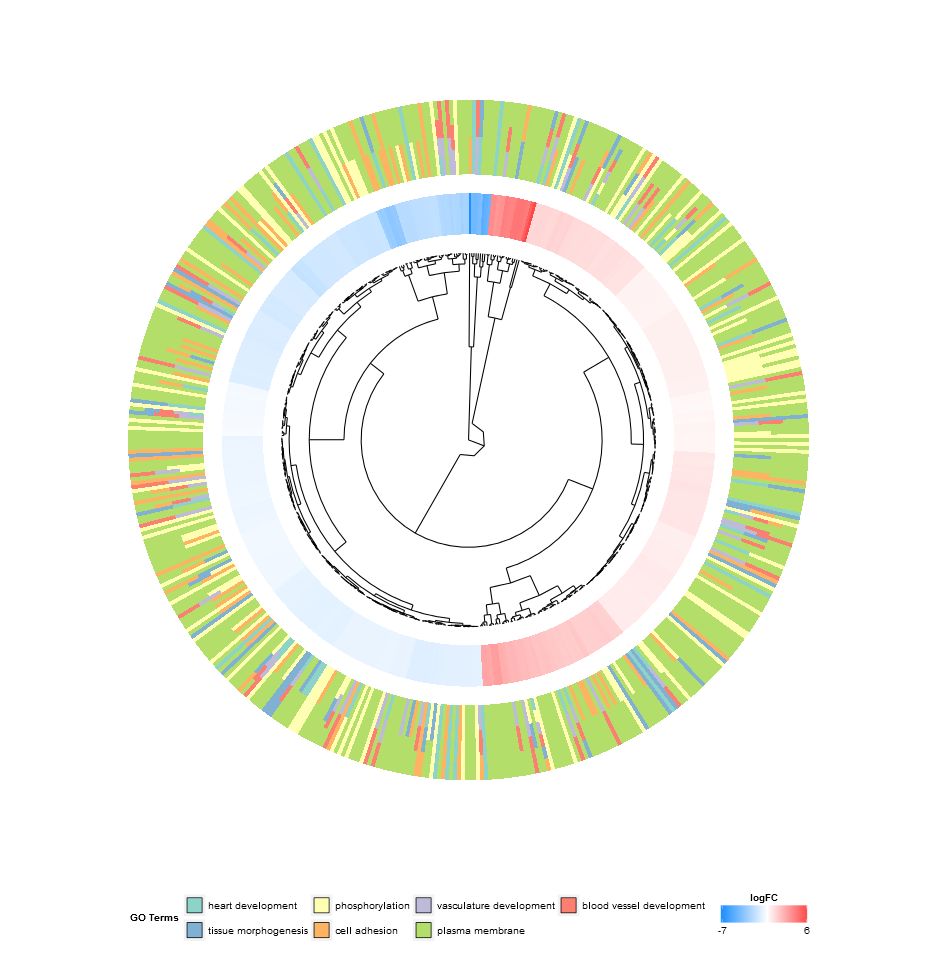
聚类(GOCluster)
- > GOCluster(circ, EC$process, clust.by = 'logFC', term.width = 2)
GOCluster()调用R内置函数hclust 对基因表水平达或根据功能分内进行层次聚类。
- > GOCluster(circ, EC$process, clust.by = 'logFC', term.width = 2)