过去几年间,高通量RNA测序已经取得了一些意想不到的成果,其中一些似乎是能够改写传统遗传学智慧的突破性发现。但是,这些调查结果现在受到了挑战,计算生物学家警告说,在这些数据密集型研究(data-intensive study)中,可能有一些潜在的统计学陷阱。
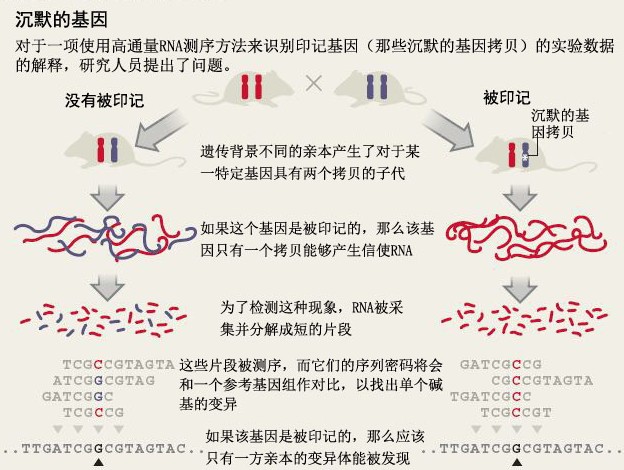
最新的一例研究是关于印记基因(imprinted gene)的。人类和其它大多数动物都会继承分别来自父方和母方的两个拷贝。但是在一些情况下,两个拷贝中只有一个能表达,另一个拷贝是沉默的。在这种情况下,我们说这个基因是“被印记的”。2010年7月,马萨诸塞州剑桥市(Cambridge,Massachusetts)哈佛大学(Harvard University)的Catherine Dulac和Christopher Gregg领导的一个研究团队在《科学》(Science)杂志上发表了一项研究,他们发现1300个小鼠基因是具有基因印记的,这个数量级比以往我们所想象的要大。
现在研究人员们正在讨论,这项研究中有缺陷的分析是否导致Catherine Dulac和Christopher Gregg在论文中错误地估计了印记基因的数量。加利福利亚州(California)斯坦福大学(Stanford University)的计算生物学家Tomas Babak指出,这项研究之所以可以发表在《科学》杂志上,是因为他们声称观察到了比前人所观察到的大一个数量级的印记基因,但是Babak并不认为这是真的。他在3月29日发表的一篇论文中公开质疑这项研究。
Dulac则表示,她和她的研究团队“毫无疑问地忠实于这些数据”,并且强调说,她们已经通过其它研究方法验证了这些发现。这一局面和另一项关于RNA测序研究论文的辩论很相似。在这项研究中,费城(Philadelphia)宾夕法尼亚大学(University of Pennsylvania)的Vivian Cheung的研究组报道了RNA编辑(RNA editing)的证据。RNA编辑使得那些由基因DNA转录成的RNA序列变得与它们的模板不一样,这种现象在人类基因组中广泛存在。RNA编辑现象之前也被观察到过,但是这项研究中观察到的RNA编辑的频繁程度极高,甚至直接挑战了传统的中心定律(central dogma)中关于生物体基因转录忠实于模板的观点。
其他的科学家们认为Cheung的研究结论很大程度产生于数据分析中的错误,而真实的RNA编辑的范围很可能并不比之前科学界认为的那样大。Cheung并没有对《自然》(Nature)杂志上这篇评论对她研究的质疑作出回应,她依然坚持自己的结论。
研究中,Dulac和Gregg使用了高通量的RNA测序来探索小鼠的RNA 以寻找单核苷酸多态性(SNP)——基因序列的一种单核苷酸突变。研究人员想知道,是否他们发现的每一个基因的SNP都可以追溯到一个或多个亲本。如果SNP主要由父母一方的基因拷贝来编码,那么研究小组就得出结论,认为该基因是被印记的。
但是Babak认为Dulac和Gregg使用的统计学方法在排除假阳性(false positive)方面是不够严格的。他的研究团队使用了多种方法来估计这项研究中的假阳性率——例如,通过使用一种更严格的标准,以及在饲养的小鼠都具有相同的遗传背景的情况下,估计印记基因假阳性事件偶然发生的概率,并在此基础上计算假阳性率。Babak的研究团队将他们计算的假阳性率用在Dulac和Gregg的数据中,并得出结论,在这篇原始论文中识别的基因印记例子大部分都很可能是假阳性。Dulac声辩说,Babak的分析很可能过滤掉真实的、但比较复杂的基因印记。
“最初遇到这些麻烦的论文实际问题都在于,它们的统计和分析整体上都做得并不是很仔细。”加州大学伯克利分校(University of California, Berkeley)的计算生物学家Lior Pachter说到,“而这将意味着你会得到完全错误的结论。”数年来,研究人员已经开发出了用来使DNA测序中的错误和偏向最小化的标准方法,但是对于高通量的RNA测序,这些方法依然还在开发中。
Pachter提到,另一个关键的问题在于,那些在某些领域高规格的论文可能仅仅进行了生物学专业的审查,但是却没有审查其计算学基础,他指出,在生物学界和数学统计学界文化是不同的,后者的外审专家往往研究被审查的论文好几个月,检查论文中的统计学和数学计算,并运行计算机程序来测试它们。
这次论战对于那些基于测序的研究具有的启示作用在于,在使用新方法确定比较罕见的遗传现象时需要统计学家的参与。斯坦福大学的一位基因组学家Jin Billy Li也是Cheung的那篇RNA编辑论文的评论者之一,他认为,在这类实验中,“如果你不仔细处理分析细节的话,你将因为低的信噪比(signal-to-noise ratio)而陷入麻烦。”
Dulac表示她的研究团队现在使用了不同的统计学方法来重新分析这些关于基因印记的数据,但她表示她非常有信心验证人员将会发现,即使采用新的方法,基因印记的发生频率也和她们之前报道的数据在同一个数量级。
以下是Nature上的原文介绍:
RNA studies under fire
High-profile results challenged over statistical analysis of sequence data.
Erika Check Hayden
High-throughput RNA sequencing has yielded some unexpected results in the past few years — including some that seem to rewrite conventional wisdom in genetics. But a few of those findings are now being challenged, as computational biologists warn of the statistical pitfalls that can lurk in data-intensive studies.
The latest case centres on imprinted genes. Humans and most other animals inherit two copies of most genes, one from each parent. But in some cases, only one copy is expressed; the other copy is silenced. In such cases, the gene is described as being imprinted. In July 2010, a team led by Catherine Dulac and Christopher Gregg, both then at Harvard University in Cambridge, Massachusetts, published a study1 in Science estimating that 1,300 mouse genes — an order of magnitude more than previously known — were imprinted.
Now, researchers are arguing that a flawed analysis led Dulac and Gregg to vastly overestimate imprinting in their paper. “The reason this paper was published in Scienceis that they made this big claim that they saw an order-of-magnitude more genes that are imprinted, and I don’t think that’s true,” says Tomas Babak, a computational biologist at Stanford University in California, who challenged the study in a paper2 published on 29 March.
Dulac counters that she and her team “absolutely stand by those data”, adding that they have confirmed some of their findings by other means. The situation resembles an ongoing debate over another RNA-sequencing paper3 published in 2011. In that study, Vivian Cheung of the University of Pennsylvania in Philadelphia and her colleagues reported evidence that RNA editing — which creates differences between a gene’s DNA sequence and the RNA sequence it gives rise to — is “widespread” in the human genome. RNA editing had been seen before, but the finding that it was so frequent challenges the central dogma, which holds that an organism’s genes are transcribed faithfully.
Other scientists have argued that Cheung’s results arose largely from errors in data analysis and that the true extent of RNA editing is probably no greater than previously thought4. Cheung did not respond to Nature’s request for comment on this story, but she has stood by her results.
For their study, Dulac and Gregg used high-throughput RNA sequencing to search mouse RNA for single nucleotide polymorphisms (SNPs) — one-letter variations in genetic sequence. The researchers then asked whether the SNPs they found for each gene could be traced to one or to both parents. If the SNPs were encoded mainly by one parent’s copy of the gene, the team concluded that the gene was imprinted (see‘The silence of the genes’).
But Babak says that the statistical methods Dulac and Gregg used were not rigorous enough to rule out false positives. His team used multiple methods to estimate the false discovery rate — for instance, by applying stricter criteria for what could be considered instances of imprinting and by estimating how many spurious examples of imprinting would appear by chance if mice from identical genetic backgrounds were bred together. Babak’s team then applied its false discovery rate to Dulac and Gregg’s data and concluded that most of the instances of imprinting identified in the original paper were probably false positives. Dulac counters that Babak’s analysis may be filtering out legitimate but complex instances of imprinting.
“What’s happened in the first few papers on these problems is that the statistics and analysis in general have not been done very carefully,” says Lior Pachter, a computational biologist at the University of California, Berkeley. “And that means you may get completely wrong answers.” Researchers have had many years to develop standard methods to minimize errors and biases in DNA sequencing, but such methods are still being developed for high-throughput RNA sequencing.
Pachter says that another key problem is that high-profile papers in the field may be well reviewed for their biology but not their computational foundations. “The culture is not the same in biology as it is in statistics or math, where reviewers sit with a paper for months, check the statistics and the math, and run the programs and test them,” he says.
The debate has implications for any sequencing-based study that requires statisticians to identify rare genetic phenomena using relatively new methods. “If you don’t deal with the analytical details very carefully, you’re going to get into trouble because of the low signal-to-noise ratio” in these types of experiments, says Jin Billy Li, a genomicist at Stanford University who was one of the critics of Cheung’s RNA-editing paper.
Dulac says that she and her colleagues are now using different statistical methods to reanalyse the imprinting data, but adds, “I am quite confident that we will find things that are likely to be around the same order of magnitude” as originally reported.
Reference
- Gregg, C. et al. Science 329, 643–648 (2010).
- DeVeale, B., van der Kooy, D. & Babak, T. PLoS Genet. 8, e1002600 (2012).
- Li, M. et al. Science 33, 53–58 (2011).
- Check Hayden, E. Nature http://dx.doi.org/10.1038/nature.2012.10217 (2012).



来自外部的引用