CellChat简介
CellChat的特点
全面的数据:大多数的细胞通讯分析方法通常只考虑配体/受体基因对,往往忽略了多亚基复合物受体和其他信号辅助因子。CellChat的作者人工挑选了2021个经过验证的细胞通讯关系,构建了新的细胞通讯参考数据库——CellChatDB。它不仅考虑了多亚基受体情况,还收录了其他重要的信号辅助因子:可溶性激动剂,拮抗剂,共刺激和共抑制膜结合受体。 高深的算法:CellChat在分析过程中使用了多种分析方法,不仅有常见的秩和检验、置换检验、SNN、KNN、UMAP,还有非负矩阵分解、社会网络分析、质量作用定律等不常用的方法。能灵活运用这么多方法分析数据,足以说明作者具有深厚的算法功底。 丰富的可视化结果:CellChat提供丰富且美观的可视化结果,有网络图、桑基图、热图、气泡图、散点图等多种图形。
安装CellChat
安装依赖项
- ## R环境安装NMF和ComplexHeatmap包
- devtools::install_github("renozao/NMF@devel")
- devtools::install_github("jokergoo/ComplexHeatmap")
- ## shell环境安装
- pip install umap-learn #如果报错,可使用conda安装
- #conda install umap-learn
安装CellChat
- devtools::install_github("sqjin/CellChat")
CellChat实践
数据来源
为了方便大家对比不同的分析方法,我们继续使用《单细胞分析之NicheNet》的数据。数据下载链接:https://zenodo.org/record/3531889/files/seuratObj.rds
初始分析
- library(CellChat)
- library(tidyverse)
- library(ggalluvial)
- rm(list=ls())
- options(stringsAsFactors = FALSE)
- ##提取表达矩阵和细胞分类信息
- scRNA <- readRDS(url("https://zenodo.org/record/3531889/files/seuratObj.rds"))
- scRNA <- UpdateSeuratObject(scRNA)
- # CellChat要求输入标准化后的表达数据
- data.input <- GetAssayData(scRNA, assay = "RNA", slot = "data")
- identity <- subset(scRNA@meta.data, select = "celltype")
- ##创建cellchat对象
- cellchat <- createCellChat(data = data.input)
- cellchat <- addMeta(cellchat, meta = identity, meta.name = "labels")
- cellchat <- setIdent(cellchat, ident.use = "labels")
- groupSize <- as.numeric(table(cellchat@idents)) # 后面有用
- ##设置参考数据库
- # 选择合适的物种,可选CellChatDB.human, CellChatDB.mouse
- CellChatDB <- CellChatDB.mouse
- # 使用"Secreted Signaling"用于细胞通讯分析
- CellChatDB.use <- subsetDB(CellChatDB, search = "Secreted Signaling")
- # 将数据库传递给cellchat对象
- cellchat@DB <- CellChatDB.use
- ##配体-受体分析
- # 提取数据库支持的数据子集
- cellchat <- subsetData(cellchat)
- # 识别过表达基因
- cellchat <- identifyOverExpressedGenes(cellchat)
- # 识别配体-受体对
- cellchat <- identifyOverExpressedInteractions(cellchat)
- # 将配体、受体投射到PPI网络
- cellchat <- projectData(cellchat, PPI.mouse)
推断细胞通讯网络
- ##推测细胞通讯网络
- cellchat <- computeCommunProb(cellchat)
- cellchat <- computeCommunProbPathway(cellchat)
- cellchat <- aggregateNet(cellchat)
细胞通讯网络系统分析及可视化
- levels(cellchat@idents) #查看细胞顺序
- vertex.receiver = c(3, 6) #指定靶细胞的索引
- cellchat@netP$pathways #查看富集到的信号通路
- pathways.show <- "CCL" #指定需要展示的通路
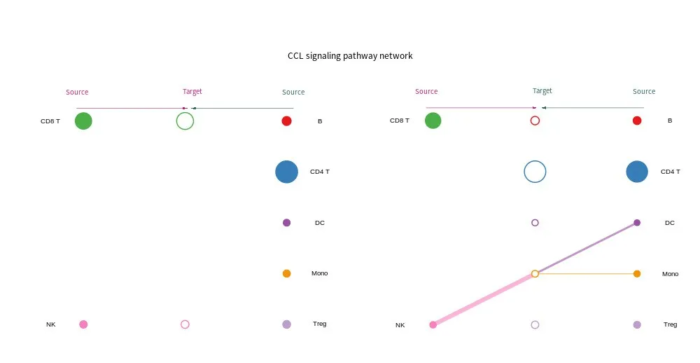
- # Hierarchy plot
- png(filename = "sig_pathway_hierarchy.png", width = 1000, height = 650)
- netVisual_aggregate(cellchat, signaling = pathways.show, vertex.receiver = vertex.receiver, vertex.size = groupSize)
- dev.off()
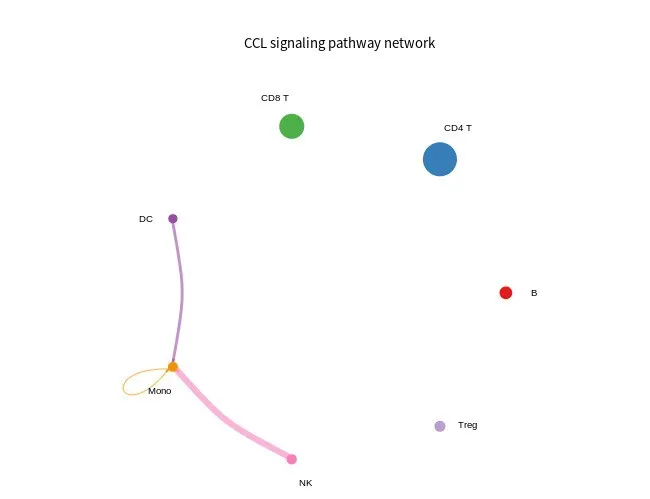
- # Circle plot
- png(filename = "sig_pathway_cricle.png", width = 650, height = 600)
- netVisual_aggregate(cellchat, signaling = pathways.show, layout = "circle", vertex.size = groupSize)
- dev.off()
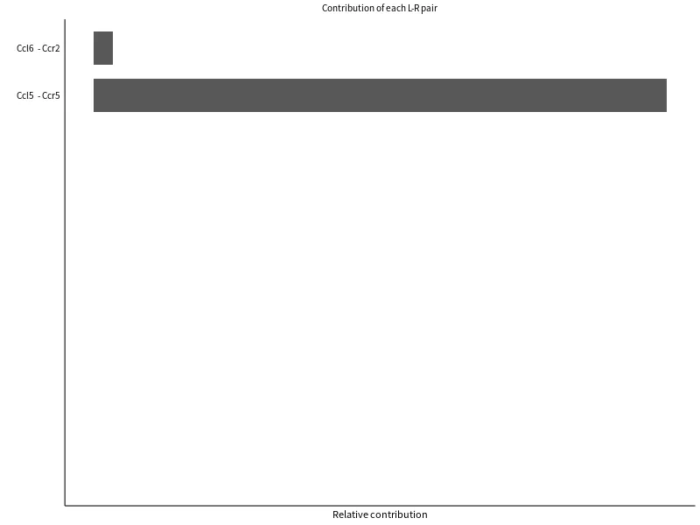
- # 计算配体-受体对信号网络的贡献度
- png(filename = "sig_pathway_L-R.png", width = 800, height = 600)
- netAnalysis_contribution(cellchat, signaling = pathways.show)
- dev.off()
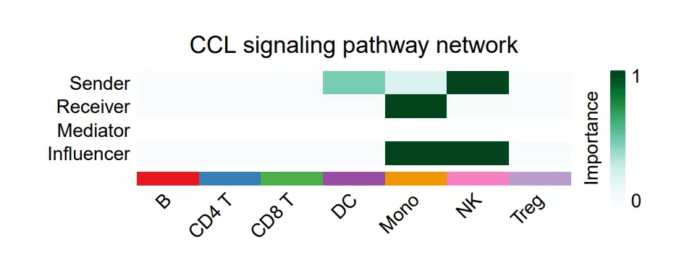
- # 分析细胞在信号网络中角色
- cellchat <- netAnalysis_signalingRole(cellchat, slot.name = "netP")
- png(filename = "sig_pathway_role.png", width = 800, height = 600)
- netVisual_signalingRole(cellchat, signaling = pathways.show)
- dev.off()
sig_pathway_hierarchy
sig_pathway_cricle
sig_pathway_L-R
sig_pathway_role
- ##细胞通讯模式和信号网络
- nPatterns = 5 #默认为5
- cellchat <- identifyCommunicationPatterns(cellchat, pattern = "outgoing", k = nPatterns)
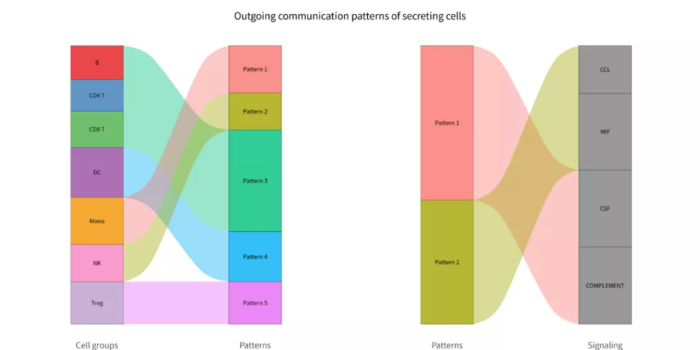
- # river plot
- p = netAnalysis_river(cellchat, pattern = "outgoing")
- ggsave("com_pattern_outgoing_river.png", p, width = 12, height = 6)
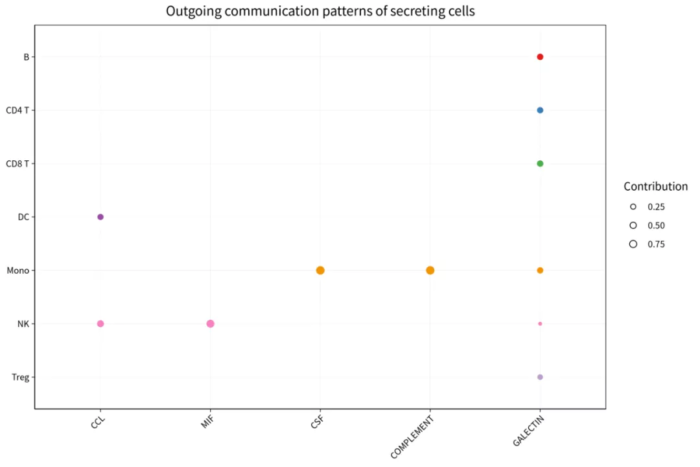
- # dot plot
- p = netAnalysis_dot(cellchat, pattern = "outgoing")
- ggsave("com_pattern_outgoing_dot.png", p, width = 9, height = 6)
- nPatterns = 5
- cellchat <- identifyCommunicationPatterns(cellchat, pattern = "incoming", k = nPatterns)
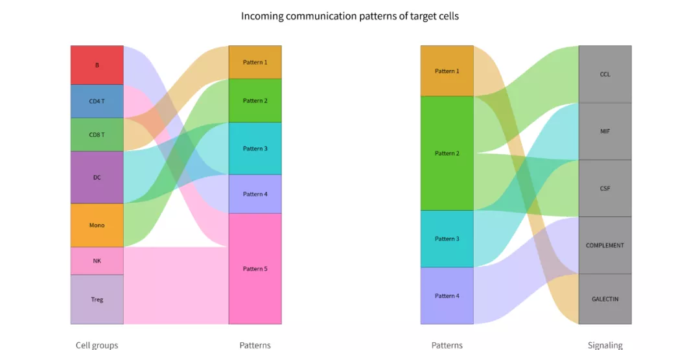
- # river plot
- p = netAnalysis_river(cellchat, pattern = "incoming")
- ggsave("com_pattern_incoming_river.png", p, width = 12, height = 6)
- # dot plot
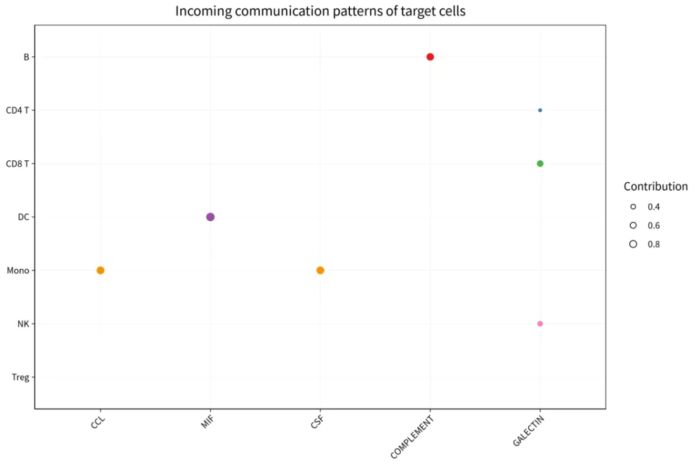
- p = netAnalysis_dot(cellchat, pattern = "incoming")
- ggsave("com_pattern_incoming_dot.png", p, width = 9, height = 6)
com_pattern_outgoing_river
com_pattern_outgoing_dot
com_pattern_incoming_river
om_pattern_incoming_dot
- ##信号网络聚类
- # 按功能相似性聚类
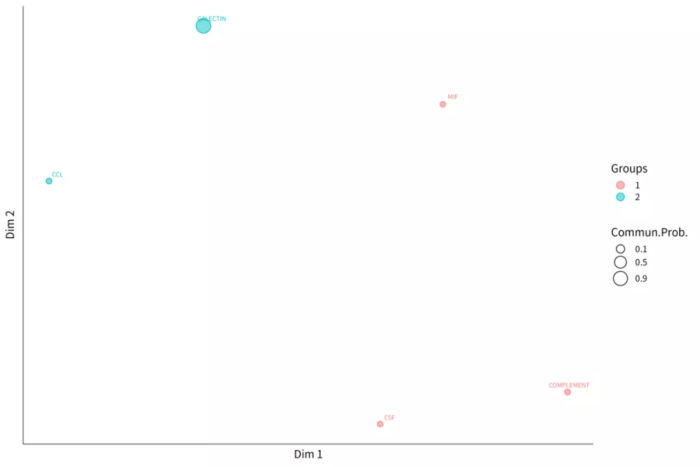
- cellchat <- computeNetSimilarity(cellchat, type = "functional")
- cellchat <- netEmbedding(cellchat, type = "functional")
- cellchat <- netClustering(cellchat, type = "functional")
- # Visualization in 2D-space
- p = netVisual_embedding(cellchat, type = "functional")
- ggsave("custer_pathway_function.png", p, width = 9, height = 6)
- p = netVisual_embeddingZoomIn(cellchat, type = "functional")
- ggsave("custer_pathway_function2.png", p, width = 8, height = 6)
- # 按结构相似性聚类
- cellchat <- computeNetSimilarity(cellchat, type = "structural")
- cellchat <- netEmbedding(cellchat, type = "structural")
- cellchat <- netClustering(cellchat, type = "structural")
- # Visualization in 2D-space
- p = netVisual_embedding(cellchat, type = "structural")
- ggsave("custer_pathway_structure.png", p, width = 9, height = 6)
- p = netVisual_embeddingZoomIn(cellchat, type = "structural")
- ggsave("custer_pathway_structure2.png", p, width = 8, height = 6)
- save(cellchat, file = "cellchat.rds")
custer_pathway_function
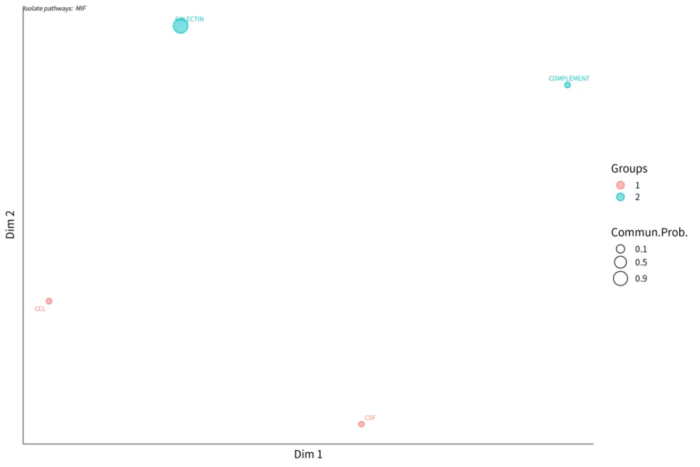
custer_pathway_structure