我们都知道,测序本身并不难,难就难在基因组的后续组装拼接,因为它涉及到大量需要考虑的问题,如重复、到位、覆盖率等等,于是如何有效的得到最后的序列或者有意义的Scaffold是做基因组面临的一个很大问题。不同的人去做会得到不同的结果,如N50、N90,scaffold数量等等。
下面简单介绍一下SOAPdenovo组装的一般过程:
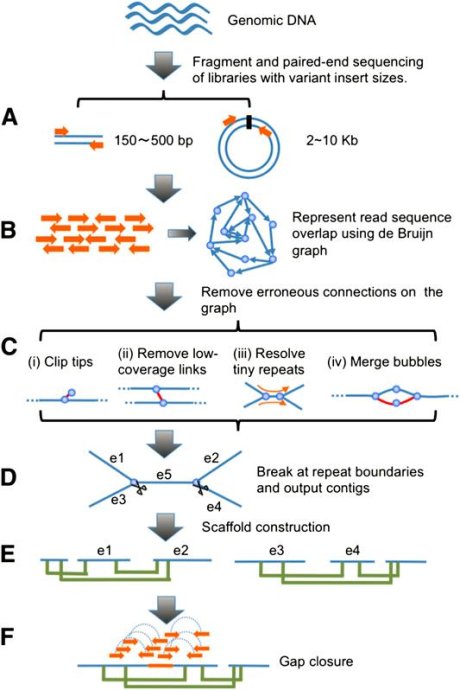
Schematic overview of the assembly algorithm.
(A) Genomic DNA was fragmented randomly and sequenced using paired-end technology.Short clones with sizes between 150 and 500 bp were amplifiedand sequenced directly; while long range (2–10 kb) paired-end libraries were constructed by circularizing DNA, fragmentation, and then purifying fragments with sizes in the range of 400–600 bp for cluster formation.
(B) The raw or precorrected reads were then loaded into computer memory and de Bruijn graph data structure was used to represent the overlap among the reads.
(C) The graph was simplified by removing erroneous connections (in red color on the graph) and solving tiny repeats by readpath:
(i) Clipping the short tips,
(ii) removing low-coverage links,(iii) solving tiny repeats by read path, and
(iv) merging the bubbles thatwere caused by repeats or heterozygotes of diploid chromosomes.
(D) On the simplified graph, we broke the connections at repeat boundaries and output the unambiguous sequence fragments as contigs.
(E)We realigned the reads onto the contigs and used the paired-end information to join the unique contigs into scaffolds.
(F) Finally, we filled in the intrascaffold gaps,which were most likely comprised by repeats, using the paired-end extracted reads.
以下是中文翻译:
A.随即打散基因组,进行双端测序:扩增长度在150--500bp之间的短克隆,并直接测序
B.将未处理(或者未经纠正的)reads读入到内存中,并且用deBruijin图数据结构来表示reads间的Overlap
C.通过移除错误的连接,解决微小的重复来简化图:
D.在简化图的基础上,我们在重复边界上打断连接,输出明确的序列作为contigs
E.我们重新用reads和contigs进行比对,使用双端信息来把单一的单一的contigs连接成scaffolds
F.最后,我们使用配对双端resds来填补scaffolds内部可能是由重复序列所造成的Gap。
更多关于SOAPdenovo算法和组装原理的介绍,请阅读原文:
本文来自:http://blog.sina.com.cn/s/blog_5d1edf6a0100w56l.html