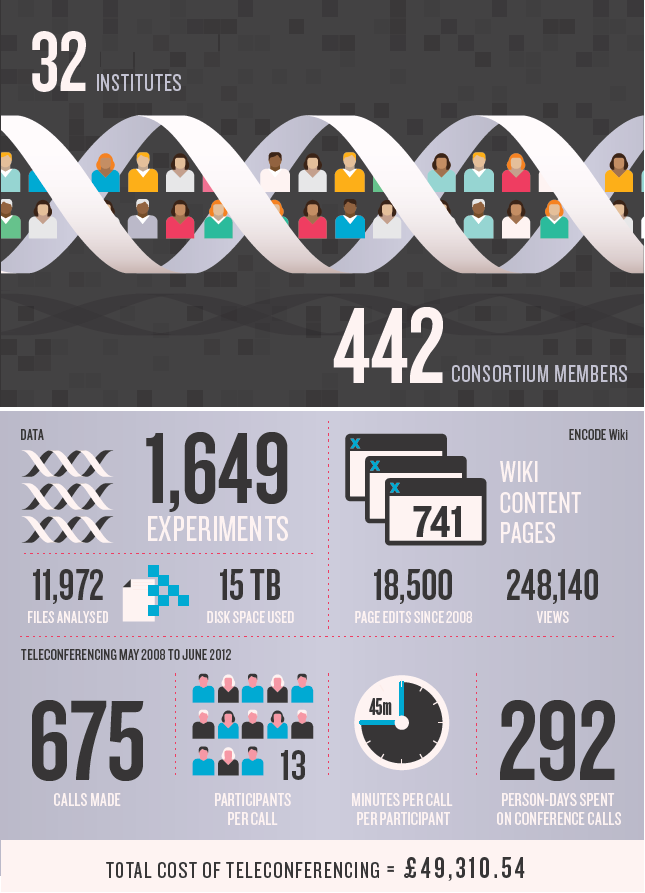
DNA元件百科全书(Encyclopedia of DNA Elements, ENCODE)项目旨在描述人类基因组中所编码的全部功能性序列元件。ENCODE计划于2003年9月正式启动,吸引了来自美国、英国、西班牙、日本和新加坡五国32个研究机构的440多名科学家的参与,经过了9年的努力,研究了147个组织类型,进行了1478次实验,获得并分析了超过15万亿字节的原始数据,确定了400万个基因开关,明确了哪些DNA片段能打开或关闭特定的基因,以及不同类型细胞之间的“开关”存在的差异。证明所谓“垃圾DNA”都是十分有用的基因成分,担任着基因调控重任。证明人体内没有一个DNA片段是无用的。
目前所有数据均全部公开(http://genome.ucsc.edu/ENCODE/),并以30篇论文在Nature、Science、Cell、JBC、Genome Biol、Genome Research同时发表(http://www.nature.com/encode)。成为一个互动的百科全书,并可以免费公开获得和利用这些全部的资料和数据。这是迄今最详细的人类基因组分析数据,是对人类生命科学的又一重大贡献。
ENCODE项目具体在以下方面取得重要进展,下面分别作简单介绍并附上相关论文的摘要与全下载链接。
1. 转录因子的足迹分析
对41种不同的细胞和组织类型进行基因组DNase I足迹分析(genomic DNase I footprinting),研究人员在DNA调节区内鉴定出4500万个转录因子结合事件,从而代表着这些转录因子与840万个不同的短DNA序列元件存在差异性地结合。他们还发现影响等位基因染色质状态的基因变异体集中分布在这些足迹之中,并且这些序列元件优先得到DNA甲基化的保护。他们鉴定出一个固定不变的50个碱基对长的足迹,并且这种足迹精确地确定着上千个人启动子内的转录起始位点。最后,他们描述了一个新的调节因子识别基序集合,其中这些基序在序列和功能上是高度保守的。
An expansive human regulatory lexicon encoded in transcription factor footprints
Regulatory factor binding to genomic DNA protects the underlying sequence from cleavage by DNase I, leaving nucleotide-resolution footprints. Using genomic DNase I footprinting across 41 diverse cell and tissue types, we detected 45 million transcription factor occupancy events within regulatory regions, representing differential binding to 8.4 million distinct short sequence elements. Here we show that this small genomic sequence compartment, roughly twice the size of the exome, encodes an expansive repertoire of conserved recognition sequences for DNA-binding proteins that nearly doubles the size of the human cis–regulatory lexicon. We find that genetic variants affecting allelic chromatin states are concentrated in footprints, and that these elements are preferentially sheltered from DNA methylation. High-resolution DNase I cleavage patterns mirror nucleotide-level evolutionary conservation and track the crystallographic topography of protein–DNA interfaces, indicating that transcription factor structure has been evolutionarily imprinted on the human genome sequence. We identify a stereotyped 50-base-pair footprint that precisely defines the site of transcript origination within thousands of human promoters. Finally, we describe a large collection of novel regulatory factor recognition motifs that are highly conserved in both sequence and function, and exhibit cell-selective occupancy patterns that closely parallel major regulators of development, differentiation and pluripotency.
论文全文下载链接:10.1038/nature11212
2. 人基因组DNA元件集成百科全书
ENCODE项目系统性地描绘出人基因组上的转录区域、转录因子结合、染色质结构和组蛋白修饰。根据这些数据,研究人员将生化功能分配到80%的人基因组,特别是在已得到很好研究的蛋白编码序列之外的区域。
An integrated encyclopedia of DNA elements in the human genome
The human genome encodes the blueprint of life, but the function of the vast majority of its nearly three billion bases is unknown. The Encyclopedia of DNA Elements (ENCODE) project has systematically mapped regions of transcription, transcription factor association, chromatin structure and histone modification. These data enabled us to assign biochemical functions for 80% of the genome, in particular outside of the well-studied protein-coding regions. Many discovered candidate regulatory elements are physically associated with one another and with expressed genes, providing new insights into the mechanisms of gene regulation. The newly identified elements also show a statistical correspondence to sequence variants linked to human disease, and can thereby guide interpretation of this variation. Overall, the project provides new insights into the organization and regulation of our genes and genome, and is an expansive resource of functional annotations for biomedical research.
论文全文下载链接:10.1038/nature11247
3. 人细胞转录全景图
RNA是基因组编码的遗传信息的直接输出。细胞的大部分调节功能都集中在RNA的合成、加工和运输、修饰和翻译之中。研究人员证实,75%的人基因组能够发生转录,并且观察到几乎所有当前已标注的RNA和上千个之前未标注的RNA的表达范围与水平、定位、加工命运、调节区和修饰。总之,这些观察结果表明人们需要重新定义基因的概念。
Landscape of transcription in human cells
Eukaryotic cells make many types of primary and processed RNAs that are found either in specific subcellular compartments or throughout the cells. A complete catalogue of these RNAs is not yet available and their characteristic subcellular localizations are also poorly understood. Because RNA represents the direct output of the genetic information encoded by genomes and a significant proportion of a cell’s regulatory capabilities are focused on its synthesis, processing, transport, modification and translation, the generation of such a catalogue is crucial for understanding genome function. Here we report evidence that three-quarters of the human genome is capable of being transcribed, as well as observations about the range and levels of expression, localization, processing fates, regulatory regions and modifications of almost all currently annotated and thousands of previously unannotated RNAs. These observations, taken together, prompt a redefinition of the concept of a gene.
论文全文下载链接:10.1038/nature11233
4. 人基因组中可访问的染色质全景图
DNase I超敏感位点(DNase I hypersensitive sites, DHSs)是调节性DNA序列的标记物。研究人员通过对125个不同的细胞和组织类型进行全基因组谱分析而鉴定出大约290万个人DHSs,并且首次大范围地绘制出人DHSs图谱。
The accessible chromatin landscape of the human genome
DNase I hypersensitive sites (DHSs) are markers of regulatory DNA and have underpinned the discovery of all classes of cis-regulatory elements including enhancers, promoters, insulators, silencers and locus control regions. Here we present the first extensive map of human DHSs identified through genome-wide profiling in 125 diverse cell and tissue types. We identify ~2.9 million DHSs that encompass virtually all known experimentally validated cis-regulatory sequences and expose a vast trove of novel elements, most with highly cell-selective regulation. Annotating these elements using ENCODE data reveals novel relationships between chromatin accessibility, transcription, DNA methylation and regulatory factor occupancy patterns. We connect ~580,000 distal DHSs with their target promoters, revealing systematic pairing of different classes of distal DHSs and specific promoter types. Patterning of chromatin accessibility at many regulatory regions is organized with dozens to hundreds of co-activated elements, and the transcellular DNase I sensitivity pattern at a given region can predict cell-type-specific functional behaviours. The DHS landscape shows signatures of recent functional evolutionary constraint. However, the DHS compartment in pluripotent and immortalized cells exhibits higher mutation rates than that in highly differentiated cells, exposing an unexpected link between chromatin accessibility, proliferative potential and patterns of human variation.
论文全文下载链接:10.1038/nature11232
5. 人基因组调控网络结构
为了确定人转录调节网络的作用原理,研究人员在450多项基因组实验中研究了119个转录相关因子的结合信息。他们发现转录因子的组合性结合是高度环境特异性的:转录因子的不同组合结合在特异性的基因组位置上。他们对所有的转录因子进行组装而产生一个层次结构,并且将它与其他基因组信息整合在一起而形成一个严密而又庞大的调节性网络。
Architecture of the human regulatory network derived from ENCODE data
Transcription factors bind in a combinatorial fashion to specify the on-and-off states of genes; the ensemble of these binding events forms a regulatory network, constituting the wiring diagram for a cell. To examine the principles of the human transcriptional regulatory network, we determined the genomic binding information of 119 transcription-related factors in over 450 distinct experiments. We found the combinatorial, co-association of transcription factors to be highly context specific: distinct combinations of factors bind at specific genomic locations. In particular, there are significant differences in the binding proximal and distal to genes. We organized all the transcription factor binding into a hierarchy and integrated it with other genomic information (for example, microRNA regulation), forming a dense meta-network. Factors at different levels have different properties; for instance, top-level transcription factors more strongly influence expression and middle-level ones co-regulate targets to mitigate information-flow bottlenecks. Moreover, these co-regulations give rise to many enriched network motifs (for example, noise-buffering feed-forward loops). Finally, more connected network components are under stronger selection and exhibit a greater degree of allele-specific activity (that is, differential binding to the two parental alleles). The regulatory information obtained in this study will be crucial for interpreting personal genome sequences and understanding basic principles of human biology and disease.
论文全文下载链接:10.1038/nature11245
6. 基因启动子的远距离相互作用全景图
在ENCODE项目中,研究人员选择1%的基因组作为项目试点区域,并且利用染色体构象捕获碳拷贝(chromosome conformation capture carbon copy, 简称为5C)技术来综合性地分析了这个区域中转录起始位点和远端序列元件之间的相互作用。他们获得GM12878、K562和HeLa-S3细胞的5C图谱。在每个细胞系,他们发现启动子和远端序列元件之间存在1000多个远距离相互作用。
The long-range interaction landscape of gene promoters
The vast non-coding portion of the human genome is full of functional elements and disease-causing regulatory variants. The principles defining the relationships between these elements and distal target genes remain unknown. Promoters and distal elements can engage in looping interactions that have been implicated in gene regulation. Here we have applied chromosome conformation capture carbon copy (5C) to interrogate comprehensively interactions between transcription start sites (TSSs) and distal elements in 1% of the human genome representing the ENCODE pilot project regions. 5C maps were generated for GM12878, K562 and HeLa-S3 cells and results were integrated with data from the ENCODE consortium. In each cell line we discovered >1,000 long-range interactions between promoters and distal sites that include elements resembling enhancers, promoters and CTCF-bound sites. We observed significant correlations between gene expression, promoter–enhancer interactions and the presence of enhancer RNAs. Long-range interactions show marked asymmetry with a bias for interactions with elements located ~120 kilobases upstream of the TSS. Long-range interactions are often not blocked by sites bound by CTCF and cohesin, indicating that many of these sites do not demarcate physically insulated gene domains. Furthermore, only ~7% of looping interactions are with the nearest gene, indicating that genomic proximity is not a simple predictor for long-range interactions. Finally, promoters and distal elements are engaged in multiple long-range interactions to form complex networks. Our results start to place genes and regulatory elements in three-dimensional context, revealing their functional relationships.
论文全文下载链接:10.1038/nature11279
7. 果蝇和人的转录因子结合位点变异分析
研究人员将ENCODE项目产生的转录因子结合图谱、他们之前发布的数据以及其他的人和果蝇等基因系中基因组变异数据来源结合在一起,来研究转录因子结合位点(transcription factor binding sites, TFBSs)的变异性。他们引入一种TFBS变异性的衡量标准和依据不断出现的每个人的转录因子结合数据来证实TFBS突变,尤其是在进化保守性位点上发生的那些突变,能够被有效地缓解从而确保转录因子结合水平保持一致性。
Analysis of variation at transcription factor binding sites inDrosophila and humans
Background
Advances in sequencing technology have boosted population genomics and made it possible to map the positions of transcription factor binding sites (TFBSs) with high precision. Here we investigate TFBS variability by combining transcription factor binding maps generated by ENCODE, modENCODE, our previously published data and other sources with genomic variation data for human individuals and Drosophila isogenic lines.
Results
We introduce a metric of TFBS variability that takes into account changes in motif match associated with mutation and makes it possible to investigate TFBS functional constraints instance-by-instance as well as in sets that share common biological properties. We also take advantage of the emerging per-individual transcription factor binding data to show evidence that TFBS mutations, particularly at evolutionarily conserved sites, can be efficiently buffered to ensure coherent levels of transcription factor binding.
Conclusions
Our analyses provide insights into the relationship between individual and interspecies variation and show evidence for the functional buffering of TFBS mutations in both humans and flies. In a broad perspective, these results demonstrate the potential of combining functional genomics and population genetics approaches for understanding gene regulation.
论文全文下载链接:10.1186/gb-2012-13-9-r49)
8. 转录因子TCF7L2通过GATA3结合到基因组上
TCF7L2转录因子与很多人类疾病相关联,如II型糖尿病和癌症。研究人员利用高通量测序技术ChIP-seq在6个人细胞系中对TCF7L2进行分析。他们鉴定出11.6万个非冗余性TCF7L2结合位点,但是只有1864 个位点在这6个细胞系中是相同的。他们还证实被H3K4me1和H3K27Ac标记的很多基因组区域也被TCF7L2结合。对细胞类型特异性的TCF7L2结合位点进行生物信息学分析揭示富集多种转录因子,包括在HepG2细胞中富集HNF4alpha和FOXA2基序,而在MCF7细胞中富集GATA3基序。转录组测序(RNA-seq)分析提示着TCF7L2通过GATA3结合到基因组上从而抑制转录。
Cell type-specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3
Background
The TCF7L2 transcription factor is linked to a variety of human diseases, including type 2 diabetes and cancer. One mechanism by which TCF7L2 could influence expression of genes involved in diverse diseases is by binding to distinct regulatory regions in different tissues. To test this hypothesis, we performed ChIP-seq for TCF7L2 in six human cell lines.
Results
We identified 116,000 non-redundant TCF7L2 binding sites, with only 1,864 sites common to the six cell lines. Using ChIP-seq, we showed that many genomic regions that are marked by both H3K4me1 and H3K27Ac are also bound by TCF7L2, suggesting that TCF7L2 plays a critical role in enhancer activity. Bioinformatic analysis of the cell type-specific TCF7L2 binding sites revealed enrichment for multiple transcription factors, including HNF4alpha and FOXA2 motifs in HepG2 cells and the GATA3 motif in MCF7 cells. ChIP-seq analysis revealed that TCF7L2 co-localizes with HNF4alpha and FOXA2 in HepG2 cells and with GATA3 in MCF7 cells. Interestingly, in MCF7 cells the TCF7L2 motif is enriched in most TCF7L2 sites but is not enriched in the sites bound by both GATA3 and TCF7L2. This analysis suggested that GATA3 might tether TCF7L2 to the genome at these sites. To test this hypothesis, we depleted GATA3 in MCF7 cells and showed that TCF7L2 binding was lost at a subset of sites. RNA-seq analysis suggested that TCF7L2 represses transcription when tethered to the genome via GATA3.
Conclusions
Our studies demonstrate a novel relationship between GATA3 and TCF7L2, and reveal important insights into TCF7L2-mediated gene regulation.
论文全文下载链接:10.1186/gb-2012-13-9-r52
9. 构建定量模型研究染色质特征和基因表达水平之间关系
通过构建出一个新的研究染色质特征和基因表达水平之间关系的定量模型,研究人员不仅证实之前在多个细胞系的研究中发现的一般性关系,而且还对它们之间的关系提出一些新的建议。Modeling gene expression using chromatin features in various cellular contexts
Background
Previous work has demonstrated that chromatin feature levels correlate with gene expression. The ENCODE project enables us to further explore this relationship using an unprecedented volume of data. Expression levels from more than 100,000 promoters were measured using a variety of high-throughput techniques applied to RNA extracted by different protocols from different cellular compartments of several human cell lines. ENCODE also generated the genome-wide mapping of eleven histone marks, one histone variant, and DNase I hypersensitivity sites in seven cell lines.
Results
We built a novel quantitative model to study the relationship between chromatin features and expression levels. Our study not only confirms that the general relationships found in previous studies hold across various cell lines, but also makes new suggestions about the relationship between chromatin features and gene expression levels. We found that expression status and expression levels can be predicted by different groups of chromatin features, both with high accuracy. We also found that expression levels measured by CAGE are better predicted than by RNA-PET or RNA-Seq, and different categories of chromatin features are the most predictive of expression for different RNA measurement methods. Additionally, PolyA+ RNA is overall more predictable than PolyA- RNA among different cell compartments, and PolyA+ cytosolic RNA measured with RNA-Seq is more predictable than PolyA+ nuclear RNA, while the opposite is true for PolyA- RNA.
Conclusions
Our study provides new insights into transcriptional regulation by analyzing chromatin features in different cellular contexts.
论文全文下载链接:10.1186/gb-2012-13-9-r53
10. GENCODE假基因资源
作为GENCODE标注人基因组的一部分,研究人员基于大规模的人工标注和计算机运算来第一次针对蛋白编码的基因进行全基因组假基因分配。他们将假基因标注和广泛性的ENCODE功能性基因组学信息整合在一起。特别的是,他们确定了每个假基因的表达水平、转录因子与RNA聚合酶II结合以及与之相关联的染色质标记。
The GENCODE pseudogene resource
Background
Pseudogenes have long been considered as nonfunctional genomic sequences. However, recent evidence suggests that many of them might have some form of biological activity, and the possibility of functionality has increased interest in their accurate annotation and integration with functional genomics data.
Results
As part of the GENCODE annotation of the human genome, we present the first genome-wide pseudogene assignment for protein-coding genes, based on both large-scale manual annotation and in silico pipelines. A key aspect of this coupled approach is that it allows us to identify pseudogenes in an unbiased fashion as well as untangle complex events through manual evaluation. We integrate the pseudogene annotations with the extensive ENCODE functional genomics information. In particular, we determine the expression level, transcription-factor and RNA polymerase II binding, and chromatin marks associated with each pseudogene. Based on their distribution, we develop simple statistical models for each type of activity, which we validate with large-scale RT-PCR-Seq experiments. Finally, we compare our pseudogenes with conservation and variation data from primate alignments and the 1000 Genomes project, producing lists of pseudogenes potentially under selection.
Conclusions
At one extreme, some pseudogenes possess conventional characteristics of functionality; these may represent genes that have recently died. On the other hand, we find interesting patterns of partial activity, which may suggest that dead genes are being resurrected as functioning non-coding RNAs. The activity data of each pseudogene are stored in an associated resource, psiDR, which will be useful for the initial identification of potentially functional pseudogenes.
论文全文下载链接:10.1186/gb-2012-13-9-r51
11. 对人启动子的转录因子结合位点进行功能性分析
为了大规模地描述转录因子结合位点功能,研究人员预测了人启动子中的455个结合位点,并对它们进行突变。在四个不同的永生化人细胞系中,他们利用瞬时转染和荧光素酶报告检测在这些位点上对主要的转录因子CTCF, GABP, GATA2, E2F, STAT和YY1进行功能性的测试。在每个细胞系中,36%到49%的结合位点提高启动子活性,并且在这些细胞系中的任何一个当中,观察到这种提高启动子活性的功能的整体发生率为70%。
Functional analysis of transcription factor binding sites in human promoters
Background
The binding of transcription factors to specific locations in the genome is integral to the orchestration of transcriptional regulation in cells. To characterize transcription factor binding site function on a large scale, we predicted and mutagenized 455 binding sites in human promoters. We carried out functional tests on these sites in four different immortalized human cell lines using transient transfections with a luciferase reporter assay, primarily for the transcription factors CTCF, GABP, GATA2, E2F, STAT, and YY1.
Results
In each cell line, between 36% and 49% of binding sites made a functional contribution to the promoter activity; the overall rate for observing function in any of the cell lines was 70%. Transcription factor binding resulted in transcriptional repression in more than a third of functional sites. When compared with predicted binding sites whose function was not experimentally verified, the functional binding sites had higher conservation and were located closer to transcriptional start sites (TSSs). Among functional sites, repressive sites tended to be located further from TSSs than were activating sites. Our data provide significant insight into the functional characteristics of YY1 binding sites, most notably the detection of distinct activating and repressing classes of YY1 binding sites. Repressing sites were located closer to, and often overlapped with, translational start sites and presented a distinctive variation on the canonical YY1 binding motif.
Conclusions
The genomic properties that we found to associate with functional TF binding sites on promoters -- conservation, TSS proximity, motifs and their variations -- point the way to improved accuracy in future TFBS predictions.
论文全文下载链接:10.1186/gb-2012-13-9-r50
12. 基于转录相关因子的结合位点对人基因组区域进行分类
研究人员通过机器学习方法构建出统计学模型来捕获三种匹配类型的区域的基因组特征:活性结合或不活性结合的区域;极端高程度共同结合区域(high degree of co-binding, HOT)和极端低程度共同结合区域(low degree of co-binding, LOT);位于基因近端或远端的调节性组件。总之,这种区域在染色体位置、染色质特征、结合到它们之上的转录因子和细胞类型特异性上存在复杂的差异。
Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors
Background
Transcription factors function by binding different classes of regulatory elements. The Encyclopedia of DNA Elements (ENCODE) project has recently produced binding data for more than 100 transcription factors from about 500 ChIP-seq experiments in multiple cell types. While this large amount of data creates a valuable resource, it is nonetheless overwhelmingly complex and simultaneously incomplete since it covers only a small fraction of all human transcription factors.
Results
As part of the consortium effort in providing a concise abstraction of the data for facilitating various types of downstream analyses, we constructed statistical models that capture the genomic features of three paired types of regions by machine-learning methods: firstly, regions with active or inactive binding; secondly, those with extremely high or low degrees of co-binding, termed HOT and LOT regions; and finally, regulatory modules proximal or distal to genes. From the distal regulatory modules, we developed computational pipelines to identify potential enhancers, many of which were validated experimentally. We further associated the predicted enhancers with potential target transcripts and the transcription factors involved. For HOT regions, we found a significant fraction of transcription factor binding without clear sequence motifs and showed that this observation could be related to strong DNA accessibility of these regions.
Conclusions
Overall, the three pairs of regions exhibit intricate differences in chromosomal locations, chromatin features, factors that bind them, and cell-type specificity. Our machine learning approach enables us to identify features potentially general to all transcription factors, including those not included in the data.
论文全文下载链接:10.1186/gb-2012-13-9-r48
13. 利用RegulomeDB标注个人基因组中的功能性变异
研究人员开发出一种新的方法和数据库,即调节物组数据库(RegulomeDB),从而能够指导人们理解人基因组中调节性序列上发生的变异。调节物组数据库包括来自ENCODE和其他来源的高通量的实验数据,以及利用计算预测和人工标注来鉴定出潜在的调节性序列变异体。
Annotation of functional variation in personal genomes using RegulomeDB
As the sequencing of healthy and disease genomes becomes more commonplace, detailed annotation provides interpretation for individual variation responsible for normal and disease phenotypes. Current approaches focus on direct changes in protein coding genes, particularly nonsynonymous mutations that directly affect the gene product. However, most individual variation occurs outside of genes and, indeed, most markers generated from genome-wide association studies (GWAS) identify variants outside of coding segments. Identification of potential regulatory changes that perturb these sites will lead to a better localization of truly functional variants and interpretation of their effects. We have developed a novel approach and database, RegulomeDB, which guides interpretation of regulatory variants in the human genome. RegulomeDB includes high-throughput, experimental data sets from ENCODE and other sources, as well as computational predictions and manual annotations to identify putative regulatory potential and identify functional variants. These data sources are combined into a powerful tool that scores variants to help separate functional variants from a large pool and provides a small set of putative sites with testable hypotheses as to their function. We demonstrate the applicability of this tool to the annotation of noncoding variants from 69 full sequenced genomes as well as that of a personal genome, where thousands of functionally associated variants were identified. Moreover, we demonstrate a GWAS where the database is able to quickly identify the known associated functional variant and provide a hypothesis as to its function. Overall, we expect this approach and resource to be valuable for the annotation of human genome sequences.
论文全文下载链接:10.1101/gr.137323.112
14. 制定ChIP-seq工作标准和指导准则
根据研究人员进行ChIP-seq实验的经历,ENCODE和modENCODE(model organism ENCODE, 模式生物ENCODE)为经常更新的ChIP-seq实验制定出一套工作标准和指导准则。
ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia
Chromatin immunoprecipitation (ChIP) followed by high-throughput DNA sequencing (ChIP-seq) has become a valuable and widely used approach for mapping the genomic location of transcription-factor binding and histone modifications in living cells. Despite its widespread use, there are considerable differences in how these experiments are conducted, how the results are scored and evaluated for quality, and how the data and metadata are archived for public use. These practices affect the quality and utility of any global ChIP experiment. Through our experience in performing ChIP-seq experiments, the ENCODE and modENCODE consortia have developed a set of working standards and guidelines for ChIP experiments that are updated routinely. The current guidelines address antibody validation, experimental replication, sequencing depth, data and metadata reporting, and data quality assessment. We discuss how ChIP quality, assessed in these ways, affects different uses of ChIP-seq data. All data sets used in the analysis have been deposited for public viewing and downloading at the ENCODE (http://encodeproject.org/ENCODE/) and modENCODE (http://www.modencode.org/) portals.
论文全文下载链接:10.1101/gr.136184.111
15. 利用RT-PCR-seq和RNA-seq统计所有人基因组编码的基因元件
在ENCODE项目中,GENCODE旨在通过人工管理和计算方法来准确地标注人基因组中所有编码蛋白的基因、假基因和非编码性的转录座位。利用一种被称作RT-PCR-seq(即先进行RT-PCR扩增,然后进行高通量多重测序)的方法可以来预测外显子连接(exon–exon junction)。研究人员验证了73%的预测结果,从而证实了1168个新的基因,其中大多数是非编码性的。
Combining RT-PCR-seq and RNA-seq to catalog all genic elements encoded in the human genome
Within the ENCODE Consortium, GENCODE aimed to accurately annotate all protein-coding genes, pseudogenes, and noncoding transcribed loci in the human genome through manual curation and computational methods. Annotated transcript structures were assessed, and less well-supported loci were systematically, experimentally validated. Predicted exon–exon junctions were evaluated by RT-PCR amplification followed by highly multiplexed sequencing readout, a method we called RT-PCR-seq. Seventy-nine percent of all assessed junctions are confirmed by this evaluation procedure, demonstrating the high quality of the GENCODE gene set. RT-PCR-seq was also efficient to screen gene models predicted using the Human Body Map (HBM) RNA-seq data. We validated 73% of these predictions, thus confirming 1168 novel genes, mostly noncoding, which will further complement the GENCODE annotation. Our novel experimental validation pipeline is extremely sensitive, far more than unbiased transcriptome profiling through RNA sequencing, which is becoming the norm. For example, exon–exon junctions unique to GENCODE annotated transcripts are five times more likely to be corroborated with our targeted approach than with extensive large human transcriptome profiling. Data sets such as the HBM and ENCODE RNA-seq data fail sampling of low-expressed transcripts. Our RT-PCR-seq targeted approach also has the advantage of identifying novel exons of known genes, as we discovered unannotated exons in ∼11% of assessed introns. We thus estimate that at least 18% of known loci have yet-unannotated exons. Our work demonstrates that the cataloging of all of the genic elements encoded in the human genome will necessitate a coordinated effort between unbiased and targeted approaches, like RNA-seq and RT-PCR-seq.
论文全文下载链接:10.1101/gr.134478.111
16. 细胞内RNA深度测序证实大多数RNA进行共转录剪接
研究人员分析了K562细胞系中通过RNA-seq测序而获得的细胞内RNA组分。他们发现在人基因组中,RNA剪接主要是在转录期间完成的。通过引入coSI 测量方法,他们证实在细胞质polyA+ RNA中,剪接几乎完全完成。因此,大多数RNA在被转录的同时进行剪接,即共转录剪接。
Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs
Splicing remains an incompletely understood process. Recent findings suggest that chromatin structure participates in its regulation. Here, we analyze the RNA from subcellular fractions obtained through RNA-seq in the cell line K562. We show that in the human genome, splicing occurs predominantly during transcription. We introduce the coSI measure, based on RNA-seq reads mapping to exon junctions and borders, to assess the degree of splicing completion around internal exons. We show that, as expected, splicing is almost fully completed in cytosolic polyA+ RNA. In chromatin-associated RNA (which includes the RNA that is being transcribed), for 5.6% of exons, the removal of the surrounding introns is fully completed, compared with 0.3% of exons for which no intron-removal has occurred. The remaining exons exist as a mixture of spliced and fewer unspliced molecules, with a median coSI of 0.75. Thus, most RNAs undergo splicing while being transcribed: “co-transcriptional splicing.” Consistent with co-transcriptional spliceosome assembly and splicing, we have found significant enrichment of spliceosomal snRNAs in chromatin-associated RNA compared with other cellular RNA fractions and other nonspliceosomal snRNAs. CoSI scores decrease along the gene, pointing to a “first transcribed, first spliced” rule, yet more downstream exons carry other characteristics, favoring rapid, co-transcriptional intron removal. Exons with low coSI values, that is, in the process of being spliced, are enriched with chromatin marks, consistent with a role for chromatin in splicing during transcription. For alternative exons and long noncoding RNAs, splicing tends to occur later, and the latter might remain unspliced in some cases.
论文全文下载链接:10.1101/gr.134445.111
17. 发现上百个小鼠和人剪接来源的miRNA
非典型的miRNA模板并不适合经常用来标注典型miRNA的策略。通过对737个小鼠和人类小RNA数据集进行大规模分析,研究人员采取严格且保守性的策略对237个小鼠剪接来源miRNA(splicing-derived miRNAs, mirtrons)和240个人mirtrons进行标注。在哺乳动物中,这些mirtrons可以分为三类:常规性的mirtrons、5'加尾mirtrons和3'加尾mirtrons。
Discovery of hundreds of mirtrons in mouse and human small RNA data
Atypical miRNA substrates do not fit criteria often used to annotate canonical miRNAs, and can escape the notice of miRNA genefinders. Recent analyses expanded the catalogs of invertebrate splicing-derived miRNAs (“mirtrons”), but only a few tens of mammalian mirtrons have been recognized to date. We performed meta-analysis of 737 mouse and human small RNA data sets comprising 2.83 billion raw reads. Using strict and conservative criteria, we provide confident annotation for 237 mouse and 240 human splicing-derived miRNAs, the vast majority of which are novel genes. These comprise three classes of splicing-derived miRNAs in mammals: conventional mirtrons, 5′-tailed mirtrons, and 3′-tailed mirtrons. In addition, we segregated several hundred additional human and mouse loci with candidate (and often compelling) evidence. Most of these loci arose relatively recently in their respective lineages. Nevertheless, some members in each of the three mirtron classes are conserved, indicating their incorporation into beneficial regulatory networks. We also provide the first Northern validation for mammalian mirtrons, and demonstrate Dicer-dependent association of mature miRNAs from all three classes of mirtrons with Ago2. The recognition of hundreds of mammalian mirtrons provides a new foundation for understanding the scope and evolutionary dynamics of Dicer substrates in mammals.
论文全文下载链接:10.1101/gr.133553.111
18. GENCODE:ENCODE项目的人基因组参照标注
GENCODE项目旨在利用计算分析、人工标注和实验验证来鉴定出人基因组中所有的基因特征。GENCODE第七版(GENCODE v7)公开发布了基因组标注数据集,包含了20687个蛋白编码的RNA基因座位、9640个长链非编码RNA基因座位,并且拥有33977个在UCSC基因数据库和RefSeq数据库中不存在的编码性转录本。它还对公开获得的长链非编码RNA(long noncoding RNA, lncRNA)进行最全面的标注。GENCODE: The reference human genome annotation for The ENCODE Project
The GENCODE Consortium aims to identify all gene features in the human genome using a combination of computational analysis, manual annotation, and experimental validation. Since the first public release of this annotation data set, few new protein-coding loci have been added, yet the number of alternative splicing transcripts annotated has steadily increased. The GENCODE 7 release contains 20,687 protein-coding and 9640 long noncoding RNA loci and has 33,977 coding transcripts not represented in UCSC genes and RefSeq. It also has the most comprehensive annotation of long noncoding RNA (lncRNA) loci publicly available with the predominant transcript form consisting of two exons. We have examined the completeness of the transcript annotation and found that 35% of transcriptional start sites are supported by CAGE clusters and 62% of protein-coding genes have annotated polyA sites. Over one-third of GENCODE protein-coding genes are supported by peptide hits derived from mass spectrometry spectra submitted to Peptide Atlas. New models derived from the Illumina Body Map 2.0 RNA-seq data identify 3689 new loci not currently in GENCODE, of which 3127 consist of two exon models indicating that they are possibly unannotated long noncoding loci. GENCODE 7 is publicly available from gencodegenes.org and via the Ensembl and UCSC Genome Browsers.
论文全文下载链接:10.1101/gr.135350.111
19. 发现人基因组中疾病相关的功能性SNP
研究人员系统性地研究了多种类型的ENCODE数据与疾病相关基因SNP(single nucleotide polymorphism, 即单核苷酸多态性)之间的关联性,并且发现在当前鉴定出的疾病关联当中,存在功能性SNP的显著性富集。
Linking disease associations with regulatory information in the human genome
Genome-wide association studies have been successful in identifying single nucleotide polymorphisms (SNPs) associated with a large number of phenotypes. However, an associated SNP is likely part of a larger region of linkage disequilibrium. This makes it difficult to precisely identify the SNPs that have a biological link with the phenotype. We have systematically investigated the association of multiple types of ENCODE data with disease-associated SNPs and show that there is significant enrichment for functional SNPs among the currently identified associations. This enrichment is strongest when integrating multiple sources of functional information and when highest confidence disease-associated SNPs are used. We propose an approach that integrates multiple types of functional data generated by the ENCODE Consortium to help identify “functional SNPs” that may be associated with the disease phenotype. Our approach generates putative functional annotations for up to 80% of all previously reported associations. We show that for most associations, the functional SNP most strongly supported by experimental evidence is a SNP in linkage disequilibrium with the reported association rather than the reported SNP itself. Our results show that the experimental data sets generated by the ENCODE Consortium can be successfully used to suggest functional hypotheses for variants associated with diseases and other phenotypes.
论文全文下载链接:10.1101/gr.136127.111
20. 在两种人细胞系中,lncRNA很少表达
ENCODE项目发现被鉴定为lncRNA的9640多个人基因组位点中,迄今为止只有大约100个得到深入的研究以便确定它们在细胞中的作用。通过共同分析ENCODE项目最近产生的两个数据集:将表达的肽链映射到它们的编码性基因组位点的串联质谱数据;ENCODE在细胞系K562和GM12878中对长polyA+和polyA-组分进行RNA-seq测序产生的数据,研究人员利用机器学习方法RuleFit3将肽链数据与RNA表达数据对应起来。他们发现大约92%的GENCODE v7发布的lncRNA在这两种细胞系中并不表达。除极少例外,核糖体能够区分编码性RNA转录本和非编码性RNA转录本,因而在lncRNA组(lncRNAome)中,异位表达和隐性mRNA都是罕见的。
Long noncoding RNAs are rarely translated in two human cell lines
Data from the Encyclopedia of DNA Elements (ENCODE) project show over 9640 human genome loci classified as long noncoding RNAs (lncRNAs), yet only ∼100 have been deeply characterized to determine their role in the cell. To measure the protein-coding output from these RNAs, we jointly analyzed two recent data sets produced in the ENCODE project: tandem mass spectrometry (MS/MS) data mapping expressed peptides to their encoding genomic loci, and RNA-seq data generated by ENCODE in long polyA+ and polyA− fractions in the cell lines K562 and GM12878. We used the machine-learning algorithm RuleFit3 to regress the peptide data against RNA expression data. The most important covariate for predicting translation was, surprisingly, the Cytosol polyA− fraction in both cell lines. LncRNAs are ∼13-fold less likely to produce detectable peptides than similar mRNAs, indicating that ∼92% of GENCODE v7 lncRNAs are not translated in these two ENCODE cell lines. Intersecting 9640 lncRNA loci with 79,333 peptides yielded 85 unique peptides matching 69 lncRNAs. Most cases were due to a coding transcript misannotated as lncRNA. Two exceptions were an unprocessed pseudogene and a bona fide lncRNA gene, both with open reading frames (ORFs) compromised by upstream stop codons. All potentially translatable lncRNA ORFs had only a single peptide match, indicating low protein abundance and/or false-positive peptide matches. We conclude that with very few exceptions, ribosomes are able to distinguish coding from noncoding transcripts and, hence, that ectopic translation and cryptic mRNAs are rare in the human lncRNAome.
论文全文下载链接:10.1101/gr.134767.111
21. 关于个人和群体的基因组调节性序列变异的基因组学
为了更好地界定人基因组调节性序列变异的模式,研究人员选择了来自不同地理位置的53个人的全基因组序列,将他们的138个细胞和组织类型的DNase I超敏感位点(DNase I hypersensitive sites, DHSs)标记的全基因组调节性DNA序列图谱结合起来。研究人员估计相比于蛋白编码的DNA序列,每个人可能拥有很多更加具有功能重要性的调节性DNA序列变异体,尽管平均而言,它们可能产生更加小的影响。
Personal and population genomics of human regulatory variation
The characteristics and evolutionary forces acting on regulatory variation in humans remains elusive because of the difficulty in defining functionally important noncoding DNA. Here, we combine genome-scale maps of regulatory DNA marked by DNase I hypersensitive sites (DHSs) from 138 cell and tissue types with whole-genome sequences of 53 geographically diverse individuals in order to better delimit the patterns of regulatory variation in humans. We estimate that individuals likely harbor many more functionally important variants in regulatory DNA compared with protein-coding regions, although they are likely to have, on average, smaller effect sizes. Moreover, we demonstrate that there is significant heterogeneity in the level of functional constraint in regulatory DNA among different cell types. We also find marked variability in functional constraint among transcription factor motifs in regulatory DNA, with sequence motifs for major developmental regulators, such as HOX proteins, exhibiting levels of constraint comparable to protein-coding regions. Finally, we perform a genome-wide scan of recent positive selection and identify hundreds of novel substrates of adaptive regulatory evolution that are enriched for biologically interesting pathways such as melanogenesis and adipocytokine signaling. These data and results provide new insights into patterns of regulatory variation in individuals and populations and demonstrate that a large proportion of functionally important variation lies beyond the exome.
论文全文下载链接:10.1101/gr.134890.111
22. 利用开放构象染色质区域来预测细胞类型特异性的基因表达
研究人员利用来自19项不同的人细胞类型的DNase-seq数据来鉴定全基因组范围的近端和远端调节性序列元件。通过匹配表达数据,他们将基因分为三类:细胞特异性的上调表达的基因、细胞特异性的下调表达的基因和组成性表达的基因。总之,他们成功地利用开放构象染色质的信息来解决利用调节性序列直接预测哺乳动物细胞特异性表达时存在的问题。
Predicting cell-type–specific gene expression from regions of open chromatin
Complex patterns of cell-type–specific gene expression are thought to be achieved by combinatorial binding of transcription factors (TFs) to sequence elements in regulatory regions. Predicting cell-type–specific expression in mammals has been hindered by the oftentimes unknown location of distal regulatory regions. To alleviate this bottleneck, we used DNase-seq data from 19 diverse human cell types to identify proximal and distal regulatory elements at genome-wide scale. Matched expression data allowed us to separate genes into classes of cell-type–specific up-regulated, down-regulated, and constitutively expressed genes. CG dinucleotide content and DNA accessibility in the promoters of these three classes of genes displayed substantial differences, highlighting the importance of including these aspects in modeling gene expression. We associated DNase I hypersensitive sites (DHSs) with genes, and trained classifiers for different expression patterns. TF sequence motif matches in DHSs provided a strong performance improvement in predicting gene expression over the typical baseline approach of using proximal promoter sequences. In particular, we achieved competitive performance when discriminating up-regulated genes from different cell types or genes up- and down-regulated under the same conditions. We identified previously known and new candidate cell-type–specific regulators. The models generated testable predictions of activating or repressive functions of regulators. DNase I footprints for these regulators were indicative of their direct binding to DNA. In summary, we successfully used information of open chromatin obtained by a single assay, DNase-seq, to address the problem of predicting cell-type–specific gene expression in mammalian organisms directly from regulatory sequence.
论文全文下载链接:10.1101/gr.135129.111
23. 探究ENCODE人RNA-seq数据中的RNA编辑
研究人员分析了来自ENCODE项目对14个人细胞系开展研究所获得的长串RNA-seq数据(这些数据经过PolyA选择,没有形成双链,且经过深度测序)以便鉴定出潜在的RNA编辑事件。他们发现RNA编辑和特异性的基因之间存在较强的关联。
RNA editing in the human ENCODE RNA-seq data
RNA-seq data can be mined for sequence differences relative to the reference genome to identify both genomic SNPs and RNA editing events. We analyzed the long, polyA-selected, unstranded, deeply sequenced RNA-seq data from the ENCODE Project across 14 human cell lines for candidate RNA editing events. On average, 43% of the RNA sequencing variants that are not in dbSNP and are within gene boundaries are A-to-G(I) RNA editing candidates. The vast majority of A-to-G(I) edits are located in introns and 3′ UTRs, with only 123 located in protein-coding sequence. In contrast, the majority of non–A-to-G variants (60%–80%) map near exon boundaries and have the characteristics of splice-mapping artifacts. After filtering out all candidates with evidence of private genomic variation using genome resequencing or ChIP-seq data, we find that up to 85% of the high-confidence RNA variants are A-to-G(I) editing candidates. Genes with A-to-G(I) edits are enriched in Gene Ontology terms involving cell division, viral defense, and translation. The distribution and character of the remaining non–A-to-G variants closely resemble known SNPs. We find no reproducible A-to-G(I) edits that result in nonsynonymous substitutions in all three lymphoblastoid cell lines in our study, unlike RNA editing in the brain. Given that only a fraction of sites are reproducibly edited in multiple cell lines and that we find a stronger association of editing and specific genes suggests that the editing of the transcript is more important than the editing of any individual site.
论文全文下载链接:10.1101/gr.134957.111
24. 细胞类型特异性的转录因子结合的序列和染色质决定簇
为了研究DNA序列信号、组蛋白修饰和DNase对细胞类型特异性的结合位点的可访问性所发挥的作用,研究人员分析了ENCODE项目所开展的286项ChIP-seq实验。与之前的研究相一致的是,他们发现DNase可访问性能够解释很多转录因子的细胞类型特异性结合。不过根据他们建立的模型,他们还发现10个转录因子拥有显著性的细胞类型特异性的结合模式,4个转录因子表现出显著不同的细胞类型特异性的DNA序列偏好性。
Sequence and chromatin determinants of cell-type–specific transcription factor binding
Gene regulatory programs in distinct cell types are maintained in large part through the cell-type–specific binding of transcription factors (TFs). The determinants of TF binding include direct DNA sequence preferences, DNA sequence preferences of cofactors, and the local cell-dependent chromatin context. To explore the contribution of DNA sequence signal, histone modifications, and DNase accessibility to cell-type–specific binding, we analyzed 286 ChIP-seq experiments performed by the ENCODE Consortium. This analysis included experiments for 67 transcriptional regulators, 15 of which were profiled in both the GM12878 (lymphoblastoid) and K562 (erythroleukemic) human hematopoietic cell lines. To model TF-bound regions, we trained support vector machines (SVMs) that use flexible k-mer patterns to capture DNA sequence signals more accurately than traditional motif approaches. In addition, we trained SVM spatial chromatin signatures to model local histone modifications and DNase accessibility, obtaining significantly more accurate TF occupancy predictions than simpler approaches. Consistent with previous studies, we find that DNase accessibility can explain cell-line–specific binding for many factors. However, we also find that of the 10 factors with prominent cell-type–specific binding patterns, four display distinct cell-type–specific DNA sequence preferences according to our models. Moreover, for two factors we identify cell-specific binding sites that are accessible in both cell types but bound only in one. For these sites, cell-type–specific sequence models, rather than DNase accessibility, are better able to explain differential binding. Our results suggest that using a single motif for each TF and filtering for chromatin accessible loci is not always sufficient to accurately account for cell-type–specific binding profiles.
论文全文下载链接:10.1101/gr.127712.111
25. 119个人转录因子结合的基因组区域附近的序列特征和染色质结构
通过对ENCODE项目在研究119个人转录因子时所获得的大约457个ChIP-seq数据集进行整合分析,研究人员在大多数数据集中鉴定出高度富集的序列基序,揭示出新的基序和验证已知的基序。
Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors
Chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) has become the dominant technique for mapping transcription factor (TF) binding regions genome-wide. We performed an integrative analysis centered around 457 ChIP-seq data sets on 119 human TFs generated by the ENCODE Consortium. We identified highly enriched sequence motifs in most data sets, revealing new motifs and validating known ones. The motif sites (TF binding sites) are highly conserved evolutionarily and show distinct footprints upon DNase I digestion. We frequently detected secondary motifs in addition to the canonical motifs of the TFs, indicating tethered binding and cobinding between multiple TFs. We observed significant position and orientation preferences between many cobinding TFs. Genes specifically expressed in a cell line are often associated with a greater occurrence of nearby TF binding in that cell line. We observed cell-line–specific secondary motifs that mediate the binding of the histone deacetylase HDAC2 and the enhancer-binding protein EP300. TF binding sites are located in GC-rich, nucleosome-depleted, and DNase I sensitive regions, flanked by well-positioned nucleosomes, and many of these features show cell type specificity. The GC-richness may be beneficial for regulating TF binding because, when unoccupied by a TF, these regions are occupied by nucleosomes in vivo. We present the results of our analysis in a TF-centric web repository Factorbook (http://factorbook.org) and will continually update this repository as more ENCODE data are generated.
论文全文下载链接:10.1101/gr.139105.112
26. 分析人lncRNA的基因结构、进化和表达
研究人员分析了迄今为止最为完整的由GENCODE项目产生的人lncRNA标注:人工标注了产生14990个RNA转录本的9277个基因。他们的分析结果表明lncRNA是通过类似于蛋白编码基因的转录途径而被产生的。而且通过在多种人器官和大脑区域所开展的lncRNA综合性表达分析,他们发现相对于蛋白编码的基因,lncRNA通常较低地表达。
The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression
The human genome contains many thousands of long noncoding RNAs (lncRNAs). While several studies have demonstrated compelling biological and disease roles for individual examples, analytical and experimental approaches to investigate these genes have been hampered by the lack of comprehensive lncRNA annotation. Here, we present and analyze the most complete human lncRNA annotation to date, produced by the GENCODE consortium within the framework of the ENCODE project and comprising 9277 manually annotated genes producing 14,880 transcripts. Our analyses indicate that lncRNAs are generated through pathways similar to that of protein-coding genes, with similar histone-modification profiles, splicing signals, and exon/intron lengths. In contrast to protein-coding genes, however, lncRNAs display a striking bias toward two-exon transcripts, they are predominantly localized in the chromatin and nucleus, and a fraction appear to be preferentially processed into small RNAs. They are under stronger selective pressure than neutrally evolving sequences—particularly in their promoter regions, which display levels of selection comparable to protein-coding genes. Importantly, about one-third seem to have arisen within the primate lineage. Comprehensive analysis of their expression in multiple human organs and brain regions shows that lncRNAs are generally lower expressed than protein-coding genes, and display more tissue-specific expression patterns, with a large fraction of tissue-specific lncRNAs expressed in the brain. Expression correlation analysis indicates that lncRNAs show particularly striking positive correlation with the expression of antisense coding genes. This GENCODE annotation represents a valuable resource for future studies of lncRNAs.
论文全文下载链接:10.1101/gr.132159.111
27. 染色质信号存在广泛的异质性
在许多种细胞系中,研究人员将14个染色质信号(12个染色质标记、DNase和核小体定位)与119个DNA结合蛋白的结合位点相关联在一起。他们开发出一种被称作CAGT(Clustered AGgregation Tool)的方法来解释染色质标记在信号强度、形状和隐性链定位上的异质性。
Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements
Gene regulation at functional elements (e.g., enhancers, promoters, insulators) is governed by an interplay of nucleosome remodeling, histone modifications, and transcription factor binding. To enhance our understanding of gene regulation, the ENCODE Consortium has generated a wealth of ChIP-seq data on DNA-binding proteins and histone modifications. We additionally generated nucleosome positioning data on two cell lines, K562 and GM12878, by MNase digestion and high-depth sequencing. Here we relate 14 chromatin signals (12 histone marks, DNase, and nucleosome positioning) to the binding sites of 119 DNA-binding proteins across a large number of cell lines. We developed a new method for unsupervised pattern discovery, the Clustered AGgregation Tool (CAGT), which accounts for the inherent heterogeneity in signal magnitude, shape, and implicit strand orientation of chromatin marks. We applied CAGT on a total of 5084 data set pairs to obtain an exhaustive catalog of high-resolution patterns of histone modifications and nucleosome positioning signals around bound transcription factors. Our analyses reveal extensive heterogeneity in how histone modifications are deposited, and how nucleosomes are positioned around binding sites. With the exception of the CTCF/cohesin complex, asymmetry of nucleosome positioning is predominant. Asymmetry of histone modifications is also widespread, for all types of chromatin marks examined, including promoter, enhancer, elongation, and repressive marks. The fine-resolution signal shapes discovered by CAGT unveiled novel correlation patterns between chromatin marks, nucleosome positioning, and sequence content. Meta-analyses of the signal profiles revealed a common vocabulary of chromatin signals shared across multiple cell lines and binding proteins.
论文全文下载链接:10.1101/gr.136366.111
28. 对转录因子结合数据进行整合分析来理解转录调节
利用对ENCODE项目产生的大量数据进行统计学模型分析来研究转录因子的转录调节。研究结果揭示不同技术和RNA抽提实验程序所捕获的转录起始位点在表达水平的预测准确度上存在显著性的差异。
Understanding transcriptional regulation by integrative analysis of transcription factor binding data
Statistical models have been used to quantify the relationship between gene expression and transcription factor (TF) binding signals. Here we apply the models to the large-scale data generated by the ENCODE project to study transcriptional regulation by TFs. Our results reveal a notable difference in the prediction accuracy of expression levels of transcription start sites (TSSs) captured by different technologies and RNA extraction protocols. In general, the expression levels of TSSs with high CpG content are more predictable than those with low CpG content. For genes with alternative TSSs, the expression levels of downstream TSSs are more predictable than those of the upstream ones. Different TF categories and specific TFs vary substantially in their contributions to predicting expression. Between two cell lines, the differential expression of TSS can be precisely reflected by the difference of TF-binding signals in a quantitative manner, arguing against the conventional on-and-off model of TF binding. Finally, we explore the relationships between TF-binding signals and other chromatin features such as histone modifications and DNase hypersensitivity for determining expression. The models imply that these features regulate transcription in a highly coordinated manner.
论文全文下载链接:10.1101/gr.136838.111
29. CTCF结合的广泛可变性与DNA甲基化相关联
CTCF是一个广泛表达的调节因子。研究人员通过研究19项不同人细胞类型的ChIP-seq数据来分析CTCF的全基因组结合模式。他们观察到高度重复性的但同时可变性非常大的基因组结合全景图,表明着CTCF结合受到高度细胞选择性的调节。
Widespread plasticity in CTCF occupancy linked to DNA methylation
CTCF is a ubiquitously expressed regulator of fundamental genomic processes including transcription, intra- and interchromosomal interactions, and chromatin structure. Because of its critical role in genome function, CTCF binding patterns have long been assumed to be largely invariant across different cellular environments. Here we analyze genome-wide occupancy patterns of CTCF by ChIP-seq in 19 diverse human cell types, including normal primary cells and immortal lines. We observed highly reproducible yet surprisingly plastic genomic binding landscapes, indicative of strong cell-selective regulation of CTCF occupancy. Comparison with massively parallel bisulfite sequencing data indicates that 41% of variable CTCF binding is linked to differential DNA methylation, concentrated at two critical positions within the CTCF recognition sequence. Unexpectedly, CTCF binding patterns were markedly different in normal versus immortal cells, with the latter showing widespread disruption of CTCF binding associated with increased methylation. Strikingly, this disruption is accompanied by up-regulation of CTCF expression, with the result that both normal and immortal cells maintain the same average number of CTCF occupancy sites genome-wide. These results reveal a tight linkage between DNA methylation and the global occupancy patterns of a major sequence-specific regulatory factor.
论文全文下载链接:10.1101/gr.136101.111
30. 细胞HepG2中高度整合的转录因子PPARGC1A结合网络
PPARGC1A是一个转录共激活因子。它结合并共同激活多种转录因子来调节大多数基因的表达。在这项研究中,研究人员在经过毛喉素(forskolin)处理的HepG2细胞中描述了一种核心的PPARGC1A转录调节网络。他们利用ChIP-seq首次描绘了PPARGC1A的全基因组结合位点,并且揭示出过多表达的对应于已知和新的PPARGC1A网络成员的DNA序列基序。他们然后利用ChIP-seq构建出6个位点特异性的转录因子结合伴侣的基因表达谱。重要的是,他们发现不同的转录因子组合结合到一套不同的功能性基因上,从而有助于揭示代谢性过程和其他细胞过程的组合性调节代码。
A highly integrated and complex PPARGC1A transcription factor binding network in HepG2 cells
PPARGC1A is a transcriptional coactivator that binds to and coactivates a variety of transcription factors (TFs) to regulate the expression of target genes. PPARGC1A plays a pivotal role in regulating energy metabolism and has been implicated in several human diseases, most notably type II diabetes. Previous studies have focused on the interplay between PPARGC1A and individual TFs, but little is known about how PPARGC1A combines with all of its partners across the genome to regulate transcriptional dynamics. In this study, we describe a core PPARGC1A transcriptional regulatory network operating in HepG2 cells treated with forskolin. We first mapped the genome-wide binding sites of PPARGC1A using chromatin-IP followed by high-throughput sequencing (ChIP-seq) and uncovered overrepresented DNA sequence motifs corresponding to known and novel PPARGC1A network partners. We then profiled six of these site-specific TF partners using ChIP-seq and examined their network connectivity and combinatorial binding patterns with PPARGC1A. Our analysis revealed extensive overlap of targets including a novel link between PPARGC1A and HSF1, a TF regulating the conserved heat shock response pathway that is misregulated in diabetes. Importantly, we found that different combinations of TFs bound to distinct functional sets of genes, thereby helping to reveal the combinatorial regulatory code for metabolic and other cellular processes. In addition, the different TFs often bound near the promoters and coding regions of each other's genes suggesting an intricate network of interdependent regulation. Overall, our study provides an important framework for understanding the systems-level control of metabolic gene expression in humans.
论文全文下载链接:10.1101/gr.127761.111



1F
解压密码多少?
B1
@ nibble 什么密码?