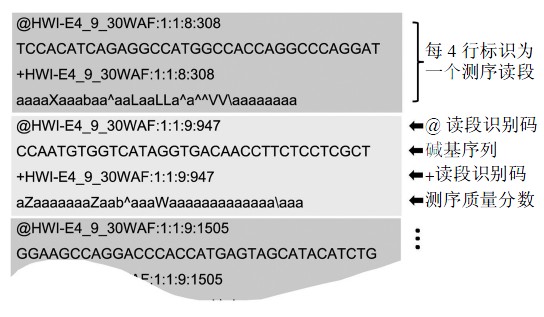
为了便于测序数据的发布和共享,高通量测序数据以FASTQ 格式来记录所测的碱基读段和质量分数.如下图 所示,FASTQ 格式以测序读段为单位存储,每条读段占4 行,其中第1 行和第3行由文件识别标志和读段名(ID)组成(第1 行以“@”开头而第3 行以“+”开头;第3 行中ID 可以省略,但“+”不能省略),第2 行为碱基序列,第4行为对应的测序质量分数.
FastQ数据格式
1.序列名称:
- 对于每一条FastQ序列,都有一个可以唯一标示的序列名称,如下:
[code lang="text"]
@HWUSI-EAS100R:6:73:941:1973#0/1
[/code]
HWUSI-EAS100R the unique instrument name 6 flowcell lane 73 tile number within the flowcell lane 941 'x'-coordinate of the cluster within the tile 1973 'y'-coordinate of the cluster within the tile #0 index number for a multiplexed sample (0 for no indexing) /1 the member of a pair, /1 or /2 (paired-end or mate-pair reads only)
Versions of the Illumina pipeline since 1.4 appear to use #NNNNNN instead of #0 for the multiplex ID, where NNNNNN is the sequence of the multiplex tag.
With Casava 1.8 the format of the '@' line has changed:
[code lang="text"]
@EAS139:136:FC706VJ:2:2104:15343:197393 1:Y:18:ATCACG
[/code]
EAS139 the unique instrument name 136 the run id FC706VJ the flowcell id 2 flowcell lane 2104 tile number within the flowcell lane 15343 'x'-coordinate of the cluster within the tile 197393 'y'-coordinate of the cluster within the tile 1 the member of a pair, 1 or 2 (paired-end or mate-pair reads only) Y Y if the read fails filter (read is bad), N otherwise 18 0 when none of the control bits are on, otherwise it is an even number ATCACG index sequence 2、质量值:对于每一条序列,其每一个碱基都有一个对应的测序质量值:
传统测序的质量值是基于Phred quality scores,定义如下:
Phred quality scores Q are defined as a property which is logarithmically related to the base-calling error probabilities P.
Q=-10 log10P
Phred quality scores are logarithmically linked to error probabilities
Phred Quality Score Probability of incorrect base call Base call accuracy 10 1 in 10 90 % 20 1 in 100 99 % 30 1 in 1000 99.9 % 40 1 in 10000 99.99 % 50 1 in 100000 99.999 % The Solexa pipeline (i.e., the software delivered with the Illumina Genome Analyzer) earlier used a different mapping, encoding the odds p/(1-p) instead of the probability p:
Although both mappings are asymptotically identical at higher quality values, they differ at lower quality levels (i.e., approximately p > 0.05, or equivalently, Q < 13).
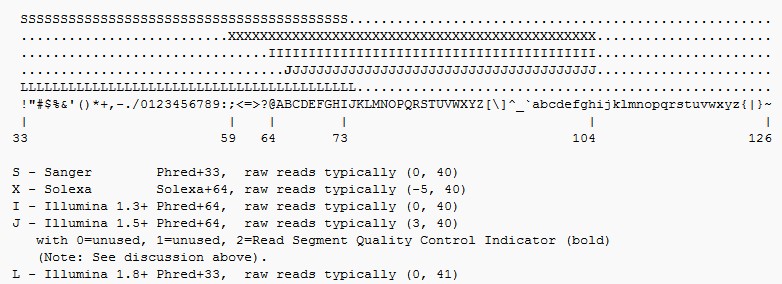
为了便于序列存储,通常采用单字符来标示序列的质量值。至于序列的quality values值,是通过一些算法得出来的。即:用字母的ASCII值减去相应的数(不同测序平台数值不一样),然后就得到Q值,然后通过前面的计算公式计算出碱基的测序错误率。
下面是不同测序平台使用的字符区间段:



来自外部的引用