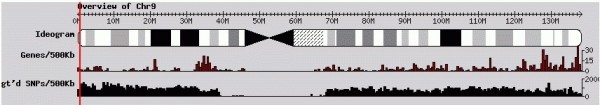
GBrowse之频率直方图,有称为频率分布图,Generating Feature Frequency Histograms,用以展示这些统计信息,可以表意以下信息:
- 不同区段内基因组Gene或者SNP等Feature区间数量分布的差异;
- 基因表达丰度;
- 序列的保守性;
新版的GBrowse更加强了该部分的功能,具体的版本是搞不清楚了,整个过程是GFF2时代使用脚本制备数据,然后再倒入数据库,然后是建立数据库的时候增加–summary参数,增加频率数据的功能,现在时默认就有,在Bio::DB::SeqFeature中,表interval_stats就是专门为支撑统计(summary)的。但这里还是要从GFF2说起,这样有利于弄清楚统计数据的GFF表示,以及如何以普通的feature方式配置进行显示,比如序列的保守性,表达丰度等,需要特殊处理,自己进行统计的生成的数据,就需要用到这个方法。
GFF2时代频率直方图
1、使用 bp_generate_histogram.pl 制备数据,脚本的用法:
- Usage: /usr/bin/bp_generate_histogram.pl [options] feature_type1 feature_type2...
- Dump out a GFF-formatted histogram of the density of the indicated set
- of feature types.
- Options:
- --dsn <dsn> Data source (default dbi:mysql:test)
- --adaptor <adaptor> Schema adaptor (default dbi::mysqlopt)
- --user <user> Username for mysql authentication
- --pass <password> Password for mysql authentication
- --bin <bp> Bin size in base pairs.
- --aggregator <list> Comma-separated list of aggregators
- --sort Sort the resulting list by type and bin
- --merge Merge features with same method but different sources
- 例如: bp_generate_histogram.pl -merge -d <> -u <> -p <> -bin 10000 SNP >snp_density.gff
2、注意生成文件的格式:
- Chr1 SNP bin 1 10000 49 + . bin Chr1:SNP
- Chr1 SNP bin 10001 20000 29 + . bin Chr1:SNP
- 注意,频率数据保存在在第六列
3、数据入库
- bp_seqfeature_load.pl -a DBI::mysql -d gb2 snp_density.gff
4、配置GBrowse
- [SNP:overview]
- feature = bin:SNP
- glyph = xyplot
- graph_type = boxes
- scale = right
- bgcolor = red
- fgcolor = red
- height = 20
- key = SNP Density
5、OK
参考:http://gmod.org/wiki/GBrowse_Configuration/Feature_frequency_histograms
新版本频率直方图的设置(Summary Mode)
bioperl、GBrowse最新的版本,使用Bio::DB::SeqFeature存储数据。只要在track设置是添加show summary参数。
- [TRACK DEFAULTS]
- ...
- show summary = 1000000
频率图xyplot的参数说明
- The following options are standard among all Glyphs. See Bio::Graphics::Glyph for a full explanation.
- Option Description Default
- ------ ----------- -------
- -fgcolor Foreground color black
- -outlinecolor Synonym for -fgcolor
- -bgcolor Background color turquoise
- -fillcolor Synonym for -bgcolor
- -linewidth Line width 1
- -height Height of glyph 10
- -font Glyph font gdSmallFont
- -label Whether to draw a label 0 (false)
- -description Whether to draw a description 0 (false)
- -hilite Highlight color undef (no color)
- In addition, the xyplot glyph recognizes the following glyph-specific options:
- Option Description Default
- ------ ----------- -------
- -max_score Maximum value of the Calculated
- feature's "score" attribute
- -min_score Minimum value of the Calculated
- feature's "score" attributes
- -graph_type Type of graph to generate. Histogram
- Options are: "histogram",
- "boxes", "line", "points",
- or "linepoints".
- -point_symbol Symbol to use. Options are none
- "triangle", "square", "disc",
- "filled_triangle",
- "filled_square",
- "filled_disc","point",
- and "none".
- -point_radius Radius of the symbol, in 4
- pixels (does not apply
- to "point")
- -scale Position where the Y axis none
- scale is drawn if any.
- It should be one of
- "left", "right", "both" or "none"
- -graph_height Specify height of the graph Same as the
- "height" option.
- -part_color For boxes & points only, none
- bgcolor of each part (should
- be a callback). Supersedes
- -neg_color.
- -scale_color Color of the scale Same as fgcolor
- -clip If min_score and/or max_score false
- are manually specified, then
- setting this to true will
- cause values outside the
- range to be clipped.
- -bicolor_pivot 0
- Where to pivot the two colors
- when drawing bicolor plots.
- Scores greater than this value will
- be drawn using -pos_color.
- Scores lower than this value will
- be drawn using -neg_color.
- -pos_color When drawing bicolor plots, same as bgcolor
- the fill color to use for
- values that are above
- the pivot point.
- -neg_color When drawing bicolor plots, same as bgcolor
- the fill color to use for values
- that are below the pivot point.
参考:
- http://gmod.org/wiki/GBrowse_2.0_HOWTO
- http://search.cpan.org/~lds/Bio-Graphics-2.21/lib/Bio/Graphics/Glyph/xyplot.pm
本文来自:http://boyun.sh.cn/bio/?p=1817