当我们进入大数据时代之后,并行运算就十分重要了。什么是大数据呢?引用一句流行语吧:
Big data is hype. Everybody talks about big data; nobody knows exactly what it is.
对于生物学来说,大数据对应的应该是第二代测序技术的兴起。所以我们谈到并行运算,主要是为了处理测序数据。在R中,有许多并行运算的包,HighPerformanceComputing就列举了当前所有的用于并行运算的软件包。为了将它更简单的呈现,我绘制了一张图,以便大家能够对常用的一些并行运算的软件包有一个大约的印象,当人们谈及这些包时,我们会有一个印象。 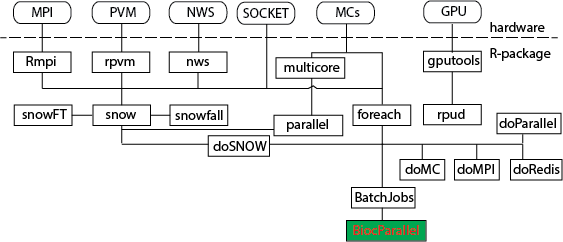 从上图中,可以看到,snow几乎关联了所有的包。但是,因为它很基础,所以很多包对它进行了再一次包装,以达到编程人员即使不了解硬件信息,也可以很轻松地移植并行运算代码的目的。而本文的重点,当然是bioconductor中最新出现的用于parallel运算的包:BiocParallel。它的优势在于它支持了很多bioconductor的类,比如GRanges数据。 下面,我们就通过一个实例来了解如何使用BiocParallel,以及在使用的过程中应该注意些什么。
从上图中,可以看到,snow几乎关联了所有的包。但是,因为它很基础,所以很多包对它进行了再一次包装,以达到编程人员即使不了解硬件信息,也可以很轻松地移植并行运算代码的目的。而本文的重点,当然是bioconductor中最新出现的用于parallel运算的包:BiocParallel。它的优势在于它支持了很多bioconductor的类,比如GRanges数据。 下面,我们就通过一个实例来了解如何使用BiocParallel,以及在使用的过程中应该注意些什么。
- > ##获取一个大一点的数据
- > library(GEOquery)
- > setwd("Downloads/")
- > getGEOSuppFiles("GSM917672")
- [1] "ftp://ftp.ncbi.nlm.nih.gov/geo/samples/GSM917nnn/GSM917672/suppl/"
- trying URL 'ftp://ftp.ncbi.nlm.nih.gov/geo/samples/GSM917nnn/GSM917672/suppl//GSM917672_286_+_density.wig.gz'
- ftp data connection made, file length 17133766 bytes
- opened URL
- ==================================================
- downloaded 16.3 Mb
- trying URL 'ftp://ftp.ncbi.nlm.nih.gov/geo/samples/GSM917nnn/GSM917672/suppl//GSM917672_286_-_density.wig.gz'
- ftp data connection made, file length 16134038 bytes
- opened URL
- ==================================================
- downloaded 15.4 Mb
- > setwd("GSM917672/")
- > sapply(dir(), gunzip)
- $`GSM917672_286_-_density.wig.gz`
- character(0)
- $`GSM917672_286_+_density.wig.gz`
- character(0)
- > file <- dir()[1]
- > ##将数据读入内存
- > s <- file.info(file)$size
- > buf <- readChar(file, s, useBytes=TRUE)
- > ##将数据按照WIG文件格式说明分割成小段,每一段只包含一段完整的WIG峰值
- > buf <- strsplit(buf, "\n", fixed=TRUE, useBytes=TRUE)[[1]]
- > infoLine <- grep("Step", buf)
- > block <- c(infoLine, length(buf)+1)
- > dif <- diff(block)
- > block <- rep(infoLine, dif)
- > buf <- split(buf, block)
- > ##WIG文件的处理
- > library(GenomicRanges)
- > parser <- function(ele){
- + library(GenomicRanges)##在并行运算中,所有的包都需要再次载入,而在非并行运算中只需要载入一次
- + firstline <- ele[1]
- + firstline <- unlist(strsplit(firstline, "\\s"))
- + firstline <- firstline[firstline!=""]
- + structure <- firstline[1]
- + firstline <- firstline[-1]
- + firstline <- do.call(rbind,strsplit(firstline, "=", fixed=TRUE))
- + firstline <- firstline[match(c("chrom","span","start","step"),
- + firstline[,1]),]
- + wiginfo <- c(structure, firstline[,2])
- + names(wiginfo) <- c("structure", "chrom","span","start","step")
- + start <- as.numeric(wiginfo["start"])
- + end <- start + 1:((length(ele)-1) * as.numeric(wiginfo["step"])) - 1
- + start <- c(start, end+1)[1:(length(ele)-1)]
- + value <- as.numeric(ele[2:length(ele)])
- + GRanges(seqnames=wiginfo["chrom"],
- + ranges=IRanges(start, end),
- + score=value)
- + }
- > ##使用pbapply中pblapply来比较运算速度。因为这个过程中也同样需要输出进度条
- > library(pbapply)
- > system.time(r1 <- pblapply(buf[1:100], parser))
- |++++++++++++++++++++++++++++++++++++++++++++++++++| 100%
- user system elapsed
- 1.474 0.017 1.512
- > system.time(r1 <- pblapply(buf[1:10000], parser))
- |++++++++++++++++++++++++++++++++++++++++++++++++++| 100%
- user system elapsed
- 152.830 0.867 157.519
- > system.time(r1 <- bplapply(buf[1:50000], parser))
- user system elapsed
- 1247.566 6.536 1257.071
- > ##使用BiocParallel来进行运算
- > library("BiocParallel")
- > library(BatchJobs)
- > cluster.functions <- makeClusterFunctionsMulticore()
- Setting for worker localhost: ncpus=8
- > bpParams <- BatchJobsParam(cluster.functions=cluster.functions)
- > system.time(r2 <- bplapply(buf[1:10000], parser, BPPARAM=bpParams))
- SubmitJobs |+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++| 100% (00:00:00)
- Waiting [S:0 R:0 D:10000 E:0] |++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++| 100% (00:00:00)0:16)
- user system elapsed
- 0.425 0.325 10.675
- > system.time(r2 <- bplapply(buf[1:10000], parser, BPPARAM=bpParams))
- SubmitJobs |+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++| 100% (00:00:00)
- Waiting [S:0 R:0 D:10000 E:0] |++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++| 100% (00:00:00)0:16)
- user system elapsed
- 7.898 2.351 166.118
- > system.time(r2 <- bplapply(buf[1:50000], parser, BPPARAM=bpParams))
- SubmitJobs |+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++| 100% (00:00:00)
- Waiting [S:0 R:0 D:50000 E:0] |++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++| 100% (00:00:00):02:51))
- user system elapsed
- 55.197 11.359 732.627
- >
- > ##比较运算结果是否一致
- > identical(r1, r2)
- [1] TRUE
从上面的步骤来看,使用BiocParallel进行运算分为三步走, 第一步,书写线程安全的程序,因为每个内核都会重新启动一个R来进行运算,所以程序的书写必须保证每个全新启动的R都可以正确的在其环境中查找到你所需要的所有变量或者函数。 第二步,设置环境参数。这一步使用makeClusterFunctions相关的函数。示例中我们使用了makeClusterFunctionsMulticore函数,其它还有makeClusterFunctionsInteractive, makeClusterFunctionsLocal, makeClusterFunctionsSSH, makeClusterFunctionsTorque, makeClusterFunctionsSGE, makeClusterFunctionsSLURM,具体可参见:?makeClusterFunctions。 第三步,使用bplapply函数提交运算。 从运算的时效性来看,如果每个单独的线程都只是很小的运算量,使用多核并不一定能节约时间,反而有可能因为等待线程结果而花费更多的时间。所以这种时候,不如使用GPU运算。如果每个单独的线程运算量很大时,使用多核就可以极大的节约时间了。 对R中的并行运算的中文博文,还可以参考:http://algosolo.com/competition/429.html 原文来自:http://pgfe.umassmed.edu/ou/archives/3396


