VEP是国际三大数据库之一的ENSEMBL提供的,也是非常主流和方便,但它是基于perl语言的,所以在模块方面可能会有点烦人。跟snpEFF一样,也是对遗传变异信息提供更具体的注释,而不仅仅是基于位点区域和基因。如果你熟悉外显子联盟这个数据库EXAC(ExAC.r0.3.sites.vep.vcf.gz),你可以下载它所有的突变记录数据,看看它对每个变异位点到底注释了些什么,它就是典型的用VEP来注释的。
随便一个位点,注释了如此多的信息!~~~
1 861389 . C T 5621.53 PASS AC=4;AC_AFR=0;AC_AMR=0;AC_Adj=4;AC_EAS=0;AC_FIN=0;AC_Het=4;AC_Hom=0;AC_NFE=3;AC_OTH=1;AC_SAS=0;AF=3.300e-05;AN=121216;AN_AFR=10212;AN_AMR=11516;AN_Adj=119730;AN_EAS=8606;AN_FIN=6594;AN_NFE=65414;AN_OTH=890;AN_SAS=16498;BaseQRankSum=2.78;ClippingRankSum=-2.380e-01;DP=1488042;FS=7.913;GQ_MEAN=62.49;GQ_STDDEV=14.73;Het_AFR=0;Het_AMR=0;Het_EAS=0;Het_FIN=0;Het_NFE=3;Het_OTH=1;Het_SAS=0;Hom_AFR=0;Hom_AMR=0;Hom_EAS=0;Hom_FIN=0;Hom_NFE=0;Hom_OTH=0;Hom_SAS=0;InbreedingCoeff=-0.0004;MQ=59.70;MQ0=0;MQRankSum=0.198;NCC=409;QD=15.11;ReadPosRankSum=0.561;VQSLOD=0.392;culprit=FS;DP_HIST=373|361|219|102|34981|16744|5493|1367|498|210|121|54|32|18|13|9|3|3|3|4,0|0|0|0|0|0|0|0|0|0|0|1|0|0|0|0|0|1|1|1;GQ_HIST=26|352|26|24|472|62|71|34|23|29|34|16|44468|8058|2176|2147|1116|370|365|739,0|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0|4;CSQ=T|ENSG00000187634|ENST00000420190|Transcript|missense_variant|157|68|23|P/L|cCg/cTg||1||1|SAMD11|HGNC|28706|protein_coding|||ENSP00000411579||Q5SV95_HUMAN&I7FV93_HUMAN&A6PWC8_HUMAN|UPI000155D47C|deleterious(0)|probably_damaging(0.999)|2/7|||ENST00000420190.1:c.68C>T|ENSP00000411579.1:p.Pro23Leu||||||||||||||||||,T|ENSG00000268179|ENST00000598827|Transcript|missense_variant|211|211|71|G/R|Ggg/Agg||1||-1|AL645608.1|Clone_based_ensembl_gene||protein_coding|YES||ENSP00000471152||M0R0C9_HUMAN|UPI0000D61E05||probably_damaging(0.98)|6/6|||ENST00000598827.1:c.211G>A|ENSP00000471152.1:p.Gly71Arg||||||||||||||||||,T|ENSG00000187634|ENST00000437963|Transcript|missense_variant|128|68|23|P/L|cCg/cTg||1||1|SAMD11|HGNC|28706|protein_coding|||ENSP00000393181||Q5SV95_HUMAN&I7FV93_HUMAN|UPI000155D47B|deleterious(0)|probably_damaging(0.999)|2/5|||ENST00000437963.1:c.68C>T|ENSP00000393181.1:p.Pro23Leu||||||||||||||||||,T|ENSG00000187634|ENST00000342066|Transcript|missense_variant|151|68|23|P/L|cCg/cTg||1||1|SAMD11|HGNC|28706|protein_coding|YES|CCDS2.2|ENSP00000342313|SAM11_HUMAN|Q5SV95_HUMAN&I7FV93_HUMAN&A6PWC8_HUMAN|UPI0000D61E04|deleterious(0)|probably_damaging(0.999)|2/14|||ENST00000342066.3:c.68C>T|ENSP00000342313.3:p.Pro23Leu||||||||||||||||||,T||ENSR00000528850|RegulatoryFeature|regulatory_region_variant|||||||1||||||regulatory_region|||||||||||||||||||||||||||||||
头文件里面有对每一列的详细介绍,包括突变的标准格式
HGVS.c –》ENST00000420190.1:c.68C>T
HGVS.p –》ENSP00000411579.1:p.Pro23Leu
还有该突变对蛋白功能的影响,包括sift和polyphen的打分~~~
不多说了,直接介绍该软件如何使用吧!
软件安装:
最新版是84:http://useast.ensembl.org/info/docs/tools/vep/script/vep_download.html
然后进入目录用perl的形式来安装这个软件:perl INSTALL.pl 即可
安装时其实有很多参数可以选择的,请仔细阅读介绍;http://useast.ensembl.org/info/docs/tools/vep/script/vep_download.html
前提是你已经安装好了两个模块!
perl -e ‘use DBD::mysql’
perl -e ‘use Archive::Extract’
如果不报错,就证明你已经安装过这些模块,如果报错,去搜索我以前关于perl模块的博客吧,不是很简单的事情。
By default the script will install the cache files in the “.vep” subdirectory of the user’s home area. Using this option users can configure where cache files are installed.
我不想把cache文件放在默认的$HOME/.vep/下面,所以我安装的时候稍微做了更改
下载完了软件,接下来就要下载注释用的数据库啦!
它支持非常多的物种的注释,我这里拿人类做例子咯:ftp://ftp.ensembl.org/pub/release-82/variation/VEP/
我下载的是ftp里面的82 版本: wget ftp://ftp.ensembl.org/pub/release-82/variation/VEP/homo_sapiens_refseq_vep_82_GRCh37.tar.gz
有6.1G,所以会有点耗时~
下载完毕后直接用tar –zxvf解压即可使用啦!
我安装软件的时候指定了cache目录,而不是默认的$HOME/.vep/
Download the archive file for your species
Extract the archive in your cache directory. By default the VEP uses $HOME/.vep/ as the cache directory, where $HOME is your UNIX home directory.
mv homo_sapiens_vep_84.tar.gz ~/.vep/ cd ~/.vep/tar xfz homo_sapiens_vep_84.tar.gz
Run the VEP with the–cache option
所以要把下载的6.1G数据库放在我自己的cashe目录
如果你安装VEP的时候用的默认安装参数,就需要把自己下载的6.1G文件放在 ~/.vep/ 目录下面
参考:http://davetang.org/wiki2/index.php?title=VEP
输入数据:
它支持好几种输入格式数据:
- BED: a simple tab-delimited format containing 3-12 columns of data. The first 3 columns contain the coordinates of the feature. If available, the VEP will use the 4th column of the file as the identifier of the feature.
- GFF: a format for describing genes and other features. If available, the VEP will use the “ID” field as the identifier of this feature.
- GTF: treated in an identical manner to GFF.
- VCF: a format used to describe genomic variants. The VEP will use the 3rd column of the file as the identifier.
- bigWig: a format for storage of dense continuous data. The VEP uses the value for the given position as the “identifier”. Note that bigWig files contain their own indices, and do not need to be indexed by tabix.
Any other files can be easily converted to be compatible with the VEP; the easiest format to produce is a BED-like file containing coordinates and an (optional) identifier:
其实重点就是给出你的突变的坐标即可,在哪条染色体,什么位置!
我们可以拿snpEFF里面的example文件夹里面的数据来做测试。
运行命令:
可以直接进入安装目录(VEP_ensembl/ensembl-tools-release-84/scripts/variant_effect_predictor)运行那个主程序
- variant_effect_predictor.pl -i example_GRCh37.vcf \
- –cache –assembly GRCh37 \
- –offline –force_overwrite
或者用全路径的形式去调用这个程序
参数非常复杂,详细介绍见:http://useast.ensembl.org/info/docs/tools/vep/script/vep_options.html
一般用标准参数就好啦,而且还有一些插件,其中我比较喜欢dbNSFP and LOFTEE plugins,这也是EXAC里面用过的。
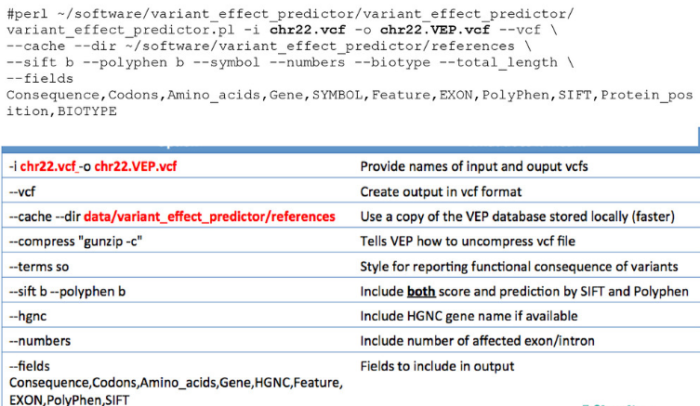
结果解读:
这个非常复杂,对结果理解了多少,就是我们对软件理解了多少。
具体大家看readme吧,注释信息太多了,按需索取:
直接看EXAC(ExAC.r0.3.sites.vep.vcf.gz)文件里面近一亿条突变记录也能慢慢理解!
参考:http://gemini.readthedocs.io/en/latest/content/functional_annotation.html
原文来自:http://www.bio-info-trainee.com/1600.html


